bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8329_orf1
Length=138
Score E
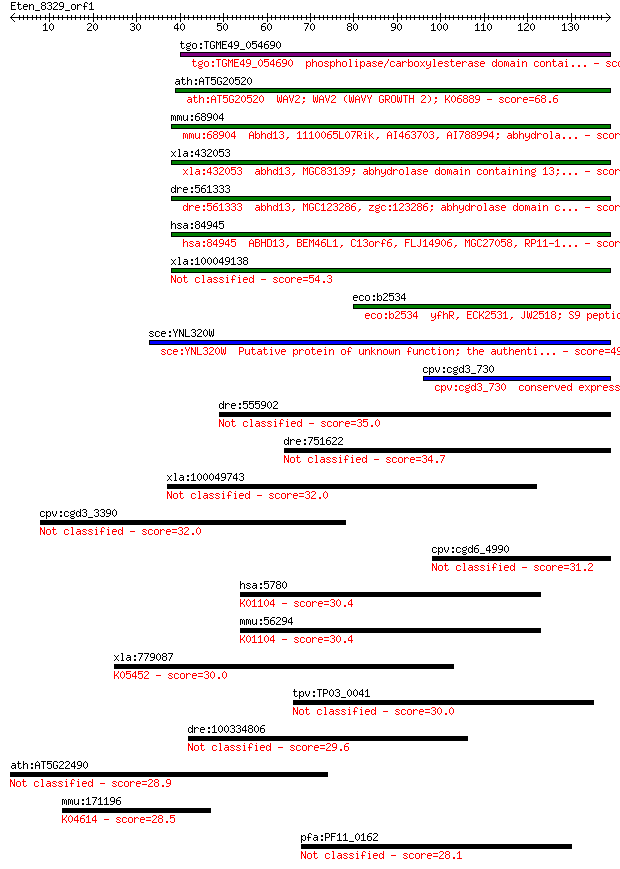
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_054690 phospholipase/carboxylesterase domain contai... 95.5 4e-20
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 68.6 6e-12
mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrola... 64.3 1e-10
xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;... 64.3 1e-10
dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain c... 62.8 3e-10
hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-1... 55.1 6e-08
xla:100049138 hypothetical protein LOC100049138 54.3 1e-07
eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein, ... 50.1 2e-06
sce:YNL320W Putative protein of unknown function; the authenti... 49.7 3e-06
cpv:cgd3_730 conserved expressed protein ; K06889 48.5 6e-06
dre:555902 Bem46-like 35.0 0.066
dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7 34.7 0.10
xla:100049743 tdp2, ad022, eap2, ttrap; tyrosyl-DNA phosphodie... 32.0 0.57
cpv:cgd3_3390 hypothetical protein 32.0 0.66
cpv:cgd6_4990 peptidase of the alpha/beta-hydrolase fold 31.2 1.0
hsa:5780 PTPN9, MEG2, PTPMEG2; protein tyrosine phosphatase, n... 30.4 1.8
mmu:56294 Ptpn9, MEG2; protein tyrosine phosphatase, non-recep... 30.4 1.9
xla:779087 gdnf-a, gdnf, gdnfa, hfb1-gdnf; glial cell derived ... 30.0 2.0
tpv:TP03_0041 hypothetical protein 30.0 2.3
dre:100334806 yeast MON (monensin-resistant) homolog family me... 29.6 2.5
ath:AT5G22490 condensation domain-containing protein 28.9 4.8
mmu:171196 Vmn1r22, V1rc23; vomeronasal 1 receptor 22; K04614 ... 28.5 6.3
pfa:PF11_0162 falcipain-3 28.1 9.0
> tgo:TGME49_054690 phospholipase/carboxylesterase domain containing
protein (EC:3.1.-.-); K06889
Length=497
Score = 95.5 bits (236), Expect = 4e-20, Method: Composition-based stats.
Identities = 48/103 (46%), Positives = 63/103 (61%), Gaps = 4/103 (3%)
Query 40 FQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENE- 98
FQEKL+F P P K P + SPA G+P+++L+L T DGV++H W +KQ +
Sbjct 47 FQEKLLFYPGVPQGFETPDKNPKGLRSPAERGLPFEELWLRTVDGVKLHCWLIKQKLPQV 106
Query 99 --KAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGY 138
APT I FHGN G+VG LP LY G+NVL++ YRGY
Sbjct 107 AAHAPTLIFFHGNAGNVGFRLPNVELLYKHVGVNVLIVSYRGY 149
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 56/101 (55%), Gaps = 7/101 (6%)
Query 39 SFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENE 98
+FQEKLV+ P+ P K +P +PA + Y+D++L + DGVR+H WF+K
Sbjct 25 AFQEKLVY---VPVLPGLSKSYPI---TPARLNLIYEDIWLQSSDGVRLHAWFIKMFPEC 78
Query 99 KAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGY 138
+ PT + F N G++ L R + + NV ++ YRGY
Sbjct 79 RGPTILFFQENAGNIAHRLEMVRIMIQKLKCNVFMLSYRGY 119
> mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrolase
domain containing 13; K06889
Length=337
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 63/104 (60%), Gaps = 11/104 (10%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDGVR++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGVRLNLILVRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGY 138
PT I FHGN G++G LP A L + +N++++DYRGY
Sbjct 110 NSPYCPTIIYFHGNAGNIGHRLPNALLMLVNLRVNLVLVDYRGY 153
> xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;
K06889
Length=336
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 63/104 (60%), Gaps = 11/104 (10%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 98 EKA--PTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGY 138
+ PT I FHGN G++G LP A L + +N++++DYRGY
Sbjct 110 NSSFSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLILVDYRGY 153
> dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain
containing 13; K06889
Length=337
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 64/104 (61%), Gaps = 11/104 (10%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQT-E 96
+ FQ+ L++ +P +P+ + + P GIP++++++ TKDG+R++ L+ T E
Sbjct 58 YKFQDVLLY---FPDQPSSSRLY-----VPMPTGIPHENVYIRTKDGIRLNLILLRYTGE 109
Query 97 N-EKAPTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGY 138
N APT + FHGN G++G +P A L + NV+++DYRGY
Sbjct 110 NPAGAPTILYFHGNAGNIGHRVPNALLMLVNLKANVVLVDYRGY 153
> hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-153I24.2,
bA153I24.2; abhydrolase domain containing 13; K06889
Length=337
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 64/104 (61%), Gaps = 11/104 (10%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDG+R++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGIRLNLILIRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGY 138
+PT I FHGN G++G LP A + + +N+L++DYRGY
Sbjct 110 NSPYSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGY 153
> xla:100049138 hypothetical protein LOC100049138
Length=336
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 63/104 (60%), Gaps = 11/104 (10%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGY 138
+PT + FHGN G++G LP A + + +N+L++DYRGY
Sbjct 110 NSNFSPTIVYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGY 153
> eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein,
function unknown; K06889
Length=284
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Query 80 TTKDGVRIHGWFLKQT---ENEKAPTFILFHGNYGHVGLTLPRARWLYDQGMNVLVIDYR 136
T KDG R+ GWF+ + + T I HGN G++ P WL ++ NV + DYR
Sbjct 55 TAKDGTRLQGWFIPSSTGPADNAIATIIHAHGNAGNMSAHWPLVSWLPERNFNVFMFDYR 114
Query 137 GY 138
G+
Sbjct 115 GF 116
> sce:YNL320W Putative protein of unknown function; the authentic,
non-tagged protein is detected in highly purified mitochondria
in high-throughput studies; K06889
Length=284
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 56/107 (52%), Gaps = 10/107 (9%)
Query 33 SIDSEFSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFL 92
S+ + + +Q +LV+ P+ + + + +P S GIPY+ L L T+D +++ W +
Sbjct 20 SVATLYHYQNRLVY-------PSWAQGARNHVDTPDSRGIPYEKLTLITQDHIKLEAWDI 72
Query 93 KQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGY 138
K EN + IL N G++G + Y Q GM+V + YRGY
Sbjct 73 KN-ENSTSTVLILCP-NAGNIGYFILIIDIFYRQFGMSVFIYSYRGY 117
> cpv:cgd3_730 conserved expressed protein ; K06889
Length=419
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query 96 ENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGY 138
+ EKAPT + FHGN G++G LPR Y+ G+N+ + YRGY
Sbjct 160 QQEKAPTIVFFHGNAGNIGHRLPRFLEFYNLIGVNIFAVSYRGY 203
> dre:555902 Bem46-like
Length=344
Score = 35.0 bits (79), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 50/116 (43%), Gaps = 26/116 (22%)
Query 49 SYPIEPAEIKKFPHIIGSPASYGIPYD-------DLFLTTKDGVRI--------HGWFLK 93
S+ + A I H++ P D +++L ++GVR+ H W
Sbjct 42 SFLVLCATIPTLQHVLSDFVDLSHPLDVGLNHTINVYLKPEEGVRVGVWHTVPEHRWKEA 101
Query 94 QTEN----EKA-----PTFILFHGNYGHVGL--TLPRARWLYDQGMNVLVIDYRGY 138
Q +N EKA P FI HGN G+ + A L G +VLV+DYRG+
Sbjct 102 QGKNAEWYEKALGDGSPIFIYLHGNGGNRSALHRIGVANVLSALGYHVLVMDYRGF 157
> dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7
Length=347
Score = 34.7 bits (78), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 45/95 (47%), Gaps = 20/95 (21%)
Query 64 IGSPASYGIPYD-DLFLTTKDGVRI--------HGWFLKQTEN----EKA-----PTFIL 105
+ P+ G+ + + +L T++GVR+ H W Q +N EKA P F+
Sbjct 65 LSRPSDLGLNHTINFYLKTEEGVRVGVWHTVPEHRWKEAQGKNVEWYEKALGDGSPIFMY 124
Query 106 FHGNYGHVGL--TLPRARWLYDQGMNVLVIDYRGY 138
HGN G+ + A L G + LV+DYRG+
Sbjct 125 LHGNTGNRSAPHRIGVANILSALGYHALVMDYRGF 159
> xla:100049743 tdp2, ad022, eap2, ttrap; tyrosyl-DNA phosphodiesterase
2 (EC:3.1.4.-)
Length=371
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 37/85 (43%), Gaps = 5/85 (5%)
Query 37 EFSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTE 96
E + Q K D + P+ K+ H + S A + DDL T + V + +KQ E
Sbjct 64 ESTLQNKPAADLADPL-----KQEMHAVTSDACIDLTSDDLVATKSEAVTSNSSTVKQQE 118
Query 97 NEKAPTFILFHGNYGHVGLTLPRAR 121
+E TF+ ++ + RAR
Sbjct 119 DESHFTFLTWNIDGLDESNVAERAR 143
> cpv:cgd3_3390 hypothetical protein
Length=1238
Score = 32.0 bits (71), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 39/76 (51%), Gaps = 12/76 (15%)
Query 8 MKMNKENLYLLFLFLLFSHFPTPSNSIDSEFSFQ---EKLVFDPSY---PIEPAEIKKFP 61
++ K NL F L S+F TP SEF F+ EKLV+ PSY I ++ F
Sbjct 257 IRSTKNNLPEACKFKLLSYFLTP-----SEFVFEILDEKLVYFPSYFRNNIISQRLEFFT 311
Query 62 HIIGSPASYGIPYDDL 77
H++ S GI Y+DL
Sbjct 312 HLM-QKQSLGIRYNDL 326
> cpv:cgd6_4990 peptidase of the alpha/beta-hydrolase fold
Length=383
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query 98 EKAPTFILFHGNYGHVGLTLPRARWLYDQGM----NVLVIDYRGY 138
EK P FI HGN +G LP W + + +VL DYR Y
Sbjct 153 EKIPVFIFSHGNATDIGSMLP---WFVNLSLKLNAHVLAYDYRSY 194
> hsa:5780 PTPN9, MEG2, PTPMEG2; protein tyrosine phosphatase,
non-receptor type 9 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=593
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Query 54 PAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTE-NEKAPTFILFHGNYGH 112
PA +KK I+G+P + +PY + L KD VR LK +E + P L G+
Sbjct 180 PARLKKV-LIVGAPIWFRVPYSIISLLLKDKVRERIQILKTSEVTQHLPRECLPENLGGY 238
Query 113 VGLTLPRARW 122
V + L A W
Sbjct 239 VKIDL--ATW 246
> mmu:56294 Ptpn9, MEG2; protein tyrosine phosphatase, non-receptor
type 9 (EC:3.1.3.48); K01104 protein-tyrosine phosphatase
[EC:3.1.3.48]
Length=593
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Query 54 PAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTE-NEKAPTFILFHGNYGH 112
PA +KK I+G+P + +PY + L KD VR LK +E + P L G+
Sbjct 180 PARLKKV-LIVGAPIWFRVPYSIISLLLKDKVRERIQILKTSEVTQHLPRECLPENLGGY 238
Query 113 VGLTLPRARW 122
V + L A W
Sbjct 239 VKIDL--ATW 246
> xla:779087 gdnf-a, gdnf, gdnfa, hfb1-gdnf; glial cell derived
neurotrophic factor; K05452 glial cell derived neurotrophic
factor
Length=227
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/78 (24%), Positives = 35/78 (44%), Gaps = 7/78 (8%)
Query 25 SHFPTPSNSIDSEFSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDG 84
SH P P D F + DP+ + P + + + I P Y +DD+ +D
Sbjct 33 SHLPDPQEGEDQVFGMDGAVPEDPTANMAPQDQQTYTEI---PDDYPDQFDDVLEFIQDT 89
Query 85 VRIHGWFLKQTENEKAPT 102
++ LK++ N++ P+
Sbjct 90 IK----RLKRSSNKQPPS 103
> tpv:TP03_0041 hypothetical protein
Length=440
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 37/73 (50%), Gaps = 9/73 (12%)
Query 66 SPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYD 125
+P S+G+ Y+ + + G W +K + N+ F+L HG +G+ TLP L
Sbjct 129 TPLSFGLSYEKV----RVGDGSESWLIK-SGNKTNKAFVLMHGWFGNPQSTLPFLNTLKQ 183
Query 126 QGM----NVLVID 134
G+ NVL+++
Sbjct 184 IGVLSDYNVLILN 196
> dre:100334806 yeast MON (monensin-resistant) homolog family
member (mon-2)-like
Length=1685
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 26/68 (38%), Positives = 33/68 (48%), Gaps = 13/68 (19%)
Query 42 EKLVFDPSYPIEPAEIKKFP----HIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
+KLVF PS P A I+ FP HI G G +DL K GV +HG +
Sbjct 1230 DKLVFVPSQPFLTALIQIFPALYQHIAG-----GFSMEDL---KKLGVILHGAVSVPISS 1281
Query 98 EKAPTFIL 105
+ +P FIL
Sbjct 1282 DSSP-FIL 1288
> ath:AT5G22490 condensation domain-containing protein
Length=482
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 18/80 (22%), Positives = 33/80 (41%), Gaps = 7/80 (8%)
Query 1 YTRSKTKMKMNKENLYLLFLFLLFSHFPTPSNSIDSEFSFQEKLVFDPSY-------PIE 53
Y + M K+N + ++ FS F + I+ +FQ K++ + + P E
Sbjct 348 YLSNVKSMIDRKKNSLITYIIYTFSEFVIKAFGINVAVAFQRKIMLNTTMCISNLPGPTE 407
Query 54 PAEIKKFPHIIGSPASYGIP 73
P +P+ YG+P
Sbjct 408 EVSFHGHPIAYFAPSIYGLP 427
> mmu:171196 Vmn1r22, V1rc23; vomeronasal 1 receptor 22; K04614
vomeronasal1 receptor
Length=302
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 13 ENLYLLF--LFLLFSHFPTPSNSIDSEFSFQEKLVF 46
N++LLF +F++ H P P + I +FSF L+F
Sbjct 18 ANMFLLFFYIFIILGHRPKPMDLILCQFSFVHILMF 53
> pfa:PF11_0162 falcipain-3
Length=492
Score = 28.1 bits (61), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 30/69 (43%), Gaps = 7/69 (10%)
Query 68 ASYGIPYDDLFLTTK-DGVRIHGWFLKQ------TENEKAPTFILFHGNYGHVGLTLPRA 120
SY +D FL + V + FLK+ T E FI+F NY + L +
Sbjct 150 VSYSNLFDTKFLMDNLETVNLFYIFLKENNKKYETSEEMQKRFIIFSENYRKIELHNKKT 209
Query 121 RWLYDQGMN 129
LY +GMN
Sbjct 210 NSLYKRGMN 218
Lambda K H
0.323 0.143 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40