bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8415_orf1
Length=88
Score E
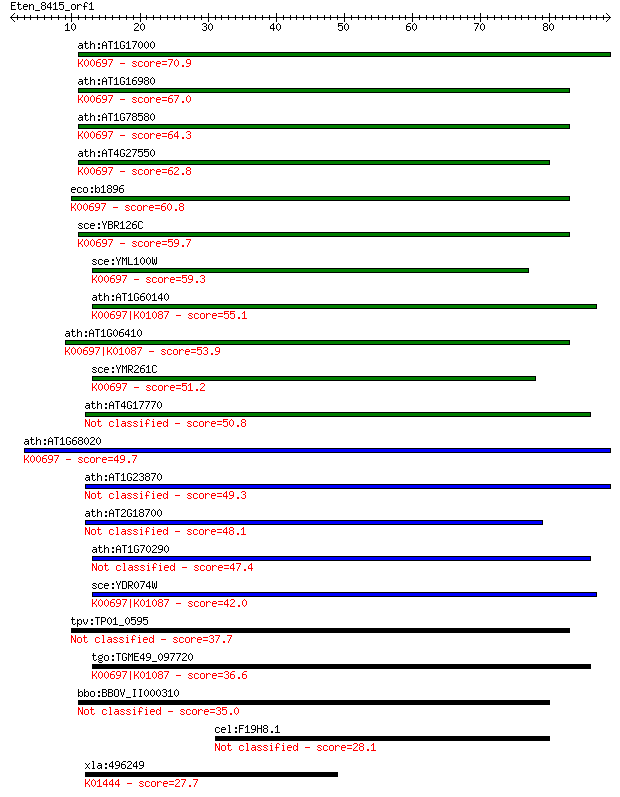
Sequences producing significant alignments: (Bits) Value
ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate ... 70.9 9e-13
ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate ... 67.0 2e-11
ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);... 64.3 8e-11
ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate ... 62.8 2e-10
eco:b1896 otsA, ECK1895, JW5312, pexA; trehalose-6-phosphate s... 60.8 1e-09
sce:YBR126C TPS1, BYP1, CIF1, FDP1, GGS1, GLC6, TSS1; Synthase... 59.7 2e-09
sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synth... 59.3 3e-09
ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transfer... 55.1 5e-08
ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate ... 53.9 1e-07
sce:YMR261C TPS3; Regulatory subunit of trehalose-6-phosphate ... 51.2 9e-07
ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase, t... 50.8 1e-06
ath:AT1G68020 ATTPS6; ATTPS6; alpha,alpha-trehalose-phosphate ... 49.7 2e-06
ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycos... 49.3 3e-06
ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups 48.1 7e-06
ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate ... 47.4 1e-05
sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the treha... 42.0 5e-04
tpv:TP01_0595 trehalose-6-phosphate synthase 37.7 0.008
tgo:TGME49_097720 trehalose-6-phosphate synthase domain-contai... 36.6 0.020
bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase do... 35.0 0.056
cel:F19H8.1 tps-2; Trehalose 6-Phosphate Synthase family membe... 28.1 6.5
xla:496249 aga; aspartylglucosaminidase (EC:3.5.1.26); K01444 ... 27.7 8.6
> ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate
synthase (UDP-forming); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=783
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
GVLILSEFAGA + L G+L++N WN + + A+ AL MP ERE RH+ + V ++
Sbjct 383 GVLILSEFAGAGQSLGAGALLVNPWNVTEVSSAIKKALNMPYEERETRHRVNFKYVKTHS 442
Query 71 STIWARKFIAALIEASDE 88
+ W F++ L +A DE
Sbjct 443 AEKWGFDFLSELNDAFDE 460
> ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=821
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 31/72 (43%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
GVL+LSEFAGA + L G+LI+N W+ + + A+ AL MP ERE RH+ Q V ++
Sbjct 405 GVLVLSEFAGAGQSLGVGALIVNPWDVTEVSSAIKEALNMPAEERETRHRSNFQYVCTHS 464
Query 71 STIWARKFIAAL 82
+ W F++ L
Sbjct 465 AEKWGLDFMSEL 476
> ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);
alpha,alpha-trehalose-phosphate synthase (UDP-forming)/ transferase,
transferring glycosyl groups; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=942
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
GVLILSEFAGAA+ L G++++N WN + A ++ AL M ERE+RH+ V +T
Sbjct 486 GVLILSEFAGAAQSLGAGAILVNPWNITEVAASIGQALNMTAEEREKRHRHNFHHVKTHT 545
Query 71 STIWARKFIAAL 82
+ WA F++ L
Sbjct 546 AQEWAETFVSEL 557
> ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=795
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 44/69 (63%), Gaps = 0/69 (0%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
GVLILSEFAGA + L G++++N WN ++ + A+ AL M E+E +HK Q V ++
Sbjct 397 GVLILSEFAGAGQSLGAGAILVNPWNIKEVSSAIGEALNMSHEEKERKHKINFQYVKTHS 456
Query 71 STIWARKFI 79
+ WA F+
Sbjct 457 TQQWADDFM 465
> eco:b1896 otsA, ECK1895, JW5312, pexA; trehalose-6-phosphate
synthase (EC:2.4.1.15); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=474
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 10 PGVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTAN 69
PGVL+LS+FAGAA L +LI+N ++ + A AL ALTM AER RH +++ V+ N
Sbjct 380 PGVLVLSQFAGAANELTS-ALIVNPYDRDEVAAALDRALTMSLAERISRHAEMLDVIVKN 438
Query 70 TSTIWARKFIAAL 82
W FI+ L
Sbjct 439 DINHWQECFISDL 451
> sce:YBR126C TPS1, BYP1, CIF1, FDP1, GGS1, GLC6, TSS1; Synthase
subunit of trehalose-6-phosphate synthase/phosphatase complex,
which synthesizes the storage carbohydrate trehalose;
also found in a monomeric form; expression is induced by the
stress response and repressed by the Ras-cAMP pathway (EC:2.4.1.15);
K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=495
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
G LILSEF GAA+ L G++I+N WN ++A+ ALT+P ++E +KL + ++ T
Sbjct 410 GSLILSEFTGAAQSLN-GAIIVNPWNTDDLSDAINEALTLPDVKKEVNWEKLYKYISKYT 468
Query 71 STIWARKFIAAL 82
S W F+ L
Sbjct 469 SAFWGENFVHEL 480
> sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synthase
(Tps1p)/phosphatase (Tps2p) complex, which converts uridine-5'-diphosphoglucose
and glucose 6-phosphate to trehalose,
homologous to Tps3p and may share function; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=1098
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
L+LSEF G+A L +G++IIN W+ + ++A+ L MP +R + KKLM+ + N ST
Sbjct 728 LLLSEFTGSASLLNDGAIIINPWDTKNFSQAILKGLEMPFDKRRPQWKKLMKDIINNDST 787
Query 73 IWAR 76
W +
Sbjct 788 NWIK 791
> ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transferase,
transferring glycosyl groups / trehalose-phosphatase;
K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]; K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=861
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 43/74 (58%), Gaps = 1/74 (1%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
L+LSEF G + L G++ +N W+ A++LY+A+TM E++ RHKK ++ +
Sbjct 476 LVLSEFIGCSPSLS-GAIRVNPWDVDAVADSLYSAITMSDFEKQLRHKKHFHYISTHDVG 534
Query 73 IWARKFIAALIEAS 86
WAR F L AS
Sbjct 535 YWARSFSQDLERAS 548
> ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=851
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/76 (36%), Positives = 45/76 (59%), Gaps = 3/76 (3%)
Query 9 GP--GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVV 66
GP +L+ SEF G + L G++ +N WN + + EAL AL+M AE++ RH+K + V
Sbjct 464 GPKKSMLVASEFIGCSPSLS-GAIRVNPWNVEATGEALNEALSMSDAEKQLRHEKHFRYV 522
Query 67 TANTSTIWARKFIAAL 82
+ + W+R F+ L
Sbjct 523 STHDVAYWSRSFLQDL 538
> sce:YMR261C TPS3; Regulatory subunit of trehalose-6-phosphate
synthase/phosphatase complex, which synthesizes the storage
carbohydrate trehalose; expression is induced by stress conditions
and repressed by the Ras-cAMP pathway; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=1054
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
L+LSEF G++ L+EG+++IN W+ A+++ +L M E+ R KKL + V + S
Sbjct 694 LLLSEFTGSSSVLKEGAILINPWDINHVAQSIKRSLEMSPEEKRRRWKKLFKSVIEHDSD 753
Query 73 IWARK 77
W K
Sbjct 754 NWITK 758
> ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase,
transferring glycosyl groups / trehalose-phosphatase
Length=862
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 42/74 (56%), Gaps = 1/74 (1%)
Query 12 VLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTS 71
+L++SEF G + L G++ +N WN EA+ AL + AE++ RH+K + V+ +
Sbjct 475 MLVVSEFIGCSPSLS-GAIRVNPWNIDAVTEAMDYALIVSEAEKQMRHEKHHKYVSTHDV 533
Query 72 TIWARKFIAALIEA 85
WAR FI L A
Sbjct 534 AYWARSFIQDLERA 547
> ath:AT1G68020 ATTPS6; ATTPS6; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=700
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 47/86 (54%), Gaps = 1/86 (1%)
Query 3 DSNLHRGPGVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKL 62
++N +L++SEF G + L G++ +N WN A+A+ +AL + E++ RH+K
Sbjct 477 EANNRNKKSMLVVSEFIGCSPSLS-GAIRVNPWNVDAVADAMDSALEVAEPEKQLRHEKH 535
Query 63 MQVVTANTSTIWARKFIAALIEASDE 88
+ V+ + WAR F+ L + E
Sbjct 536 YKYVSTHDVGYWARSFLQDLERSCGE 561
> ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=867
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query 12 VLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTS 71
+L++SEF G + L G++ +N W+ AEA+ ALTM E+ RH+K V+ +
Sbjct 475 MLVVSEFIGCSPSLS-GAIRVNPWDVDAVAEAVNLALTMGETEKRLRHEKHYHYVSTHDV 533
Query 72 TIWARKFIAALIEASDE 88
WA+ F+ L A E
Sbjct 534 GYWAKSFMQDLERACRE 550
> ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups
Length=862
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 12 VLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTS 71
V+I+SEF G + L G++ +N WN A+ +A+TM E+ RH+K + ++++
Sbjct 467 VIIVSEFIGCSPSLS-GAIRVNPWNIDAVTNAMSSAMTMSDKEKNLRHQKHHKYISSHNV 525
Query 72 TIWARKF 78
WAR +
Sbjct 526 AYWARSY 532
> ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=856
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 41/73 (56%), Gaps = 1/73 (1%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
L++SEF G + L G++ +N W+ AEA+ +AL M E++ RH+K ++ +
Sbjct 471 LVVSEFIGCSPSLS-GAIRVNPWDVDAVAEAVNSALKMSETEKQLRHEKHYHYISTHDVG 529
Query 73 IWARKFIAALIEA 85
WA+ F+ L A
Sbjct 530 YWAKSFMQDLERA 542
> sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the trehalose-6-phosphate
synthase/phosphatase complex, which synthesizes
the storage carbohydrate trehalose; expression is induced
by stress conditions and repressed by the Ras-cAMP pathway
(EC:3.1.3.12); K00697 alpha,alpha-trehalose-phosphate synthase
(UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=896
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 44/74 (59%), Gaps = 3/74 (4%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
LILSEF+G++ L++ ++++N W+ A+++ AL + + E+ KL + V T
Sbjct 473 LILSEFSGSSNVLKD-AIVVNPWDSVAVAKSINMALKLDKEEKSNLESKLWKEVP--TIQ 529
Query 73 IWARKFIAALIEAS 86
W KF+++L E +
Sbjct 530 DWTNKFLSSLKEQA 543
> tpv:TP01_0595 trehalose-6-phosphate synthase
Length=1059
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 10 PGVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTAN 69
P + I+SEF+ ++ L ++ IN W+ EAL TA++ A E KK + + N
Sbjct 637 PAIAIISEFSAFSKTLLS-AIRINPWHTYNVMEALDTAMSSDPATTLESCKKDVSYIENN 695
Query 70 TSTIWARKFIAAL 82
+ W FI L
Sbjct 696 DAVTWVNDFIKEL 708
> tgo:TGME49_097720 trehalose-6-phosphate synthase domain-containing
protein (EC:2.4.1.15 2.7.9.5 3.1.3.12); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15];
K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=1222
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 38/73 (52%), Gaps = 1/73 (1%)
Query 13 LILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTST 72
+ILSEF G + L ++ +N W + A+A+ + MP E+ +R + ++ N++
Sbjct 659 VILSEFTGCSRALAS-AIRVNPWKVEAVADAMDRIINMPVEEQRDRFTRDRDYLSHNSTQ 717
Query 73 IWARKFIAALIEA 85
WA + I L A
Sbjct 718 KWADENILDLRRA 730
> bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase
domain containing protein
Length=958
Score = 35.0 bits (79), Expect = 0.056, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 1/69 (1%)
Query 11 GVLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANT 70
G +ILSEF G ++ L ++ +N W+ + EAL A+ ER E + V ++
Sbjct 562 GSVILSEFVGFSKMLLS-AIRVNPWHIESVVEALDRAMCYSSEERAEVALHDFEYVMSSD 620
Query 71 STIWARKFI 79
+ W F+
Sbjct 621 TVAWVDHFV 629
> cel:F19H8.1 tps-2; Trehalose 6-Phosphate Synthase family member
(tps-2)
Length=1229
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 31 IINSWNEQQSAEALYTALTMPRAEREERHKKLMQVVTANTSTIWARKFI 79
I N ++ +A Y+A P R E K+L + + AN W+ F+
Sbjct 738 ISNVFDPDSYCDAFYSAALEPEDVRAEHGKRLHEFIMANDIERWSCAFL 786
> xla:496249 aga; aspartylglucosaminidase (EC:3.5.1.26); K01444
N4-(beta-N-acetylglucosaminyl)-L-asparaginase [EC:3.5.1.26]
Length=347
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 12 VLILSEFAGAAEFLQEGSLIINSWNEQQSAEALYTAL 48
VL + AAEF E L+IN+W+ + + EA + L
Sbjct 10 VLCVLTLVCAAEFNSELPLVINTWSFRNATEAAWRVL 46
Lambda K H
0.313 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40