bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8513_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
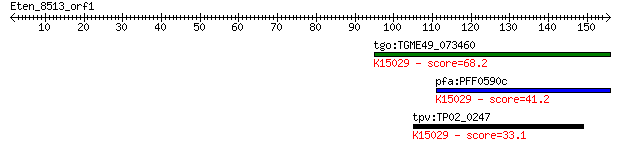
tgo:TGME49_073460 eukaryotic translation initiation factor 3 s... 68.2 1e-11
pfa:PFF0590c homologue of human HSPC025; K15029 translation in... 41.2 0.001
tpv:TP02_0247 hypothetical protein; K15029 translation initiat... 33.1 0.41
> tgo:TGME49_073460 eukaryotic translation initiation factor 3
subunit 6 interacting protein, putative ; K15029 translation
initiation factor 3 subunit L
Length=639
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 45/62 (72%), Gaps = 1/62 (1%)
Query 95 HQQRERVEAE-LADEDLDRMMAERVAFEPEVETFLRELYDKLYVRCTDSVRRLYEFDYPS 153
+Q + VE E L D LDRMM E+V FE +VE FLR LYD++Y R D++RRLYE ++P+
Sbjct 44 NQASQNVEGEGLDDAALDRMMGEKVRFENDVEEFLRSLYDEVYDRNVDAIRRLYEVEFPA 103
Query 154 LS 155
L+
Sbjct 104 LT 105
> pfa:PFF0590c homologue of human HSPC025; K15029 translation
initiation factor 3 subunit L
Length=656
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 111 DRMMAERVAFEPEVETFLRELYDKLYVRCTDSVRRLYEFDYPSLS 155
+ ++ E V+FE EVETFL L+D +Y R +++++L++ D+ ++S
Sbjct 9 ENVIEEIVSFEEEVETFLINLHDFVYHRNAEAIKKLFDTDFYTIS 53
> tpv:TP02_0247 hypothetical protein; K15029 translation initiation
factor 3 subunit L
Length=544
Score = 33.1 bits (74), Expect = 0.41, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 105 LADEDLDRMMAERVAFEPEVETFLRELYDKLYVRCTDSVRRLYE 148
LA + R + + + EPEV FL L+D LY + ++++ LYE
Sbjct 11 LATDGPQRTVTQLLPIEPEVVDFLVRLHDNLYRKNAEALKNLYE 54
Lambda K H
0.316 0.123 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40