bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8524_orf1
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
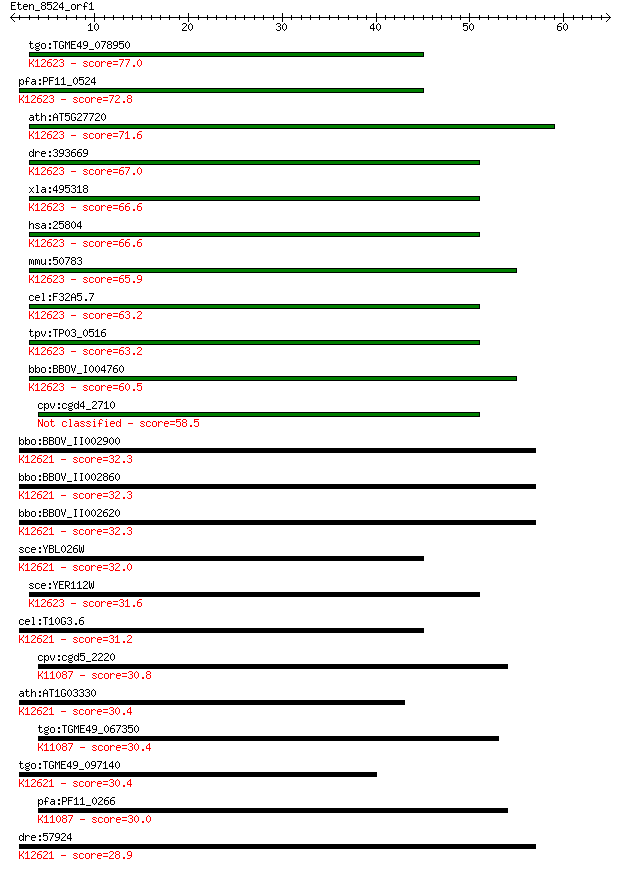
tgo:TGME49_078950 U6 snRNA associated Sm-like protein LSm4, pu... 77.0 1e-14
pfa:PF11_0524 lsm4 homologue, putative; K12623 U6 snRNA-associ... 72.8 2e-13
ath:AT5G27720 emb1644 (embryo defective 1644); K12623 U6 snRNA... 71.6 6e-13
dre:393669 lsm4, MGC73057, zgc:73057, zgc:77848; LSM4 homolog,... 67.0 2e-11
xla:495318 lsm4; LSM4 homolog, U6 small nuclear RNA associated... 66.6 2e-11
hsa:25804 LSM4, YER112W; LSM4 homolog, U6 small nuclear RNA as... 66.6 2e-11
mmu:50783 Lsm4; LSM4 homolog, U6 small nuclear RNA associated ... 65.9 3e-11
cel:F32A5.7 lsm-4; LSM Sm-like protein family member (lsm-4); ... 63.2 2e-10
tpv:TP03_0516 U6 snRNA-associated protein; K12623 U6 snRNA-ass... 63.2 2e-10
bbo:BBOV_I004760 19.m02880; U6 snRNA-associated Sm-like protei... 60.5 1e-09
cpv:cgd4_2710 hypothetical protein 58.5 5e-09
bbo:BBOV_II002900 18.m06239; u6 snRNA-associated sm-like prote... 32.3 0.35
bbo:BBOV_II002860 18.m06235; u6 snRNA-associated sm-like prote... 32.3 0.35
bbo:BBOV_II002620 18.m06211; u6 snRNA-associated sm-like prote... 32.3 0.35
sce:YBL026W LSM2, SMX5, SNP3; Lsm (Like Sm) protein; part of h... 32.0 0.56
sce:YER112W LSM4, SDB23, USS1; Lsm (Like Sm) protein; part of ... 31.6 0.72
cel:T10G3.6 gut-2; GUT differentiation defective family member... 31.2 0.90
cpv:cgd5_2220 small nuclear ribonucleoprotein D1. SM domain co... 30.8 1.2
ath:AT1G03330 small nuclear ribonucleoprotein D, putative / sn... 30.4 1.3
tgo:TGME49_067350 small nuclear ribonucleoprotein, putative ; ... 30.4 1.3
tgo:TGME49_097140 u6 snRNA-associated sm-like protein Lsm2, pu... 30.4 1.6
pfa:PF11_0266 small nuclear ribonucleoprotein D1, putative; K1... 30.0 2.1
dre:57924 smx5, lsm2, wu:fe48h10, zgc:101795; smx5; K12621 U6 ... 28.9 4.2
> tgo:TGME49_078950 U6 snRNA associated Sm-like protein LSm4,
putative ; K12623 U6 snRNA-associated Sm-like protein LSm4
Length=214
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 31/42 (73%), Positives = 40/42 (95%), Gaps = 0/42 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELV 44
FMNLHLK+SVCTSKDGERF+K++EC+IRGN +KY RLQ+E++
Sbjct 36 FMNLHLKDSVCTSKDGERFWKLSECYIRGNMVKYIRLQDEVL 77
> pfa:PF11_0524 lsm4 homologue, putative; K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=126
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 26/43 (60%), Positives = 40/43 (93%), Gaps = 0/43 (0%)
Query 2 RFMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELV 44
RFMNLH+KN +CTSKDG++F+K++EC++RGN+IKY R+Q++ +
Sbjct 35 RFMNLHMKNIICTSKDGDKFWKISECYVRGNSIKYIRVQDQAI 77
> ath:AT5G27720 emb1644 (embryo defective 1644); K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=129
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDENPKAGTSP 58
+MN+HL+ +CTSKDG+RF+++ EC+IRGNTIKY R+ +E++ V++E + P
Sbjct 35 WMNIHLREVICTSKDGDRFWRMPECYIRGNTIKYLRVPDEVIDKVQEEKTRTDRKP 90
> dre:393669 lsm4, MGC73057, zgc:73057, zgc:77848; LSM4 homolog,
U6 small nuclear RNA associated (S. cerevisiae); K12623 U6
snRNA-associated Sm-like protein LSm4
Length=143
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 42/48 (87%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN++L+ +CTS+DG++F+++ EC+IRG+TIKY R+ +E++ +VK+E
Sbjct 35 WMNINLREVICTSRDGDKFWRMPECYIRGSTIKYLRIPDEIIDMVKEE 82
> xla:495318 lsm4; LSM4 homolog, U6 small nuclear RNA associated;
K12623 U6 snRNA-associated Sm-like protein LSm4
Length=138
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 42/48 (87%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN++L+ +CTS+DG++F+++ EC+IRG+TIKY R+ +E++ +VK+E
Sbjct 35 WMNINLREVICTSRDGDKFWRMPECYIRGSTIKYLRIPDEIIDMVKEE 82
> hsa:25804 LSM4, YER112W; LSM4 homolog, U6 small nuclear RNA
associated (S. cerevisiae); K12623 U6 snRNA-associated Sm-like
protein LSm4
Length=139
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 42/48 (87%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN++L+ +CTS+DG++F+++ EC+IRG+TIKY R+ +E++ +VK+E
Sbjct 35 WMNINLREVICTSRDGDKFWRMPECYIRGSTIKYLRIPDEIIDMVKEE 82
> mmu:50783 Lsm4; LSM4 homolog, U6 small nuclear RNA associated
(S. cerevisiae); K12623 U6 snRNA-associated Sm-like protein
LSm4
Length=138
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 43/52 (82%), Gaps = 0/52 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDENPKA 54
+MN++L+ +CTS+DG++F+++ EC+IRG+TIKY R+ +E++ +V++E K
Sbjct 35 WMNINLREVICTSRDGDKFWRMPECYIRGSTIKYLRIPDEIIDMVREEAAKG 86
> cel:F32A5.7 lsm-4; LSM Sm-like protein family member (lsm-4);
K12623 U6 snRNA-associated Sm-like protein LSm4
Length=123
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/48 (54%), Positives = 39/48 (81%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MN+HL + + TSKDG++FFK++E ++RG+TIKY R+ E +V LVK E
Sbjct 36 WMNIHLVDVIFTSKDGDKFFKMSEAYVRGSTIKYLRIPETVVDLVKTE 83
> tpv:TP03_0516 U6 snRNA-associated protein; K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=141
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
FMN+H+ N VCTSK G F+K+ ECFIRGNT+K RL E+ + K+E
Sbjct 36 FMNMHMVNVVCTSKSGTDFWKLDECFIRGNTVKSLRLPNEVAEIAKEE 83
> bbo:BBOV_I004760 19.m02880; U6 snRNA-associated Sm-like protein
LSm4; K12623 U6 snRNA-associated Sm-like protein LSm4
Length=104
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/52 (53%), Positives = 38/52 (73%), Gaps = 1/52 (1%)
Query 3 FMNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDENPKA 54
FMN+H+ N++CTSK G+ F+K++ECFIRGN +K R +E VA V E KA
Sbjct 24 FMNVHMINAICTSKKGDEFWKLSECFIRGNNVKSFRFPDE-VATVALEESKA 74
> cpv:cgd4_2710 hypothetical protein
Length=77
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 4 MNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
MNL L N +C S+DG FFK+ EC+IRGN IK R+ +E + KD+
Sbjct 1 MNLALNNVICNSRDGNGFFKMVECYIRGNNIKLIRISDENINTAKDD 47
> bbo:BBOV_II002900 18.m06239; u6 snRNA-associated sm-like protein;
K12621 U6 snRNA-associated Sm-like protein LSm2
Length=99
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 2 RFMNLHLKNSVCTSKDGERF---FKVAECFIRGNTIKYARLQ------EELVALVKDENP 52
+++N L N T+ D ER+ V CF+RG+ ++Y L EEL L + E
Sbjct 36 QYLNFKLSN--VTASDTERYPHLLSVVNCFVRGSVVRYVYLNSSDVNTEELQELCRREAM 93
Query 53 KAGT 56
K+
Sbjct 94 KSNV 97
> bbo:BBOV_II002860 18.m06235; u6 snRNA-associated sm-like protein
Lsm2; K12621 U6 snRNA-associated Sm-like protein LSm2
Length=99
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 2 RFMNLHLKNSVCTSKDGERF---FKVAECFIRGNTIKYARLQ------EELVALVKDENP 52
+++N L N T+ D ER+ V CF+RG+ ++Y L EEL L + E
Sbjct 36 QYLNFKLSN--VTASDTERYPHLLSVVNCFVRGSVVRYVYLNSSDVNTEELQELCRREAM 93
Query 53 KAGT 56
K+
Sbjct 94 KSNV 97
> bbo:BBOV_II002620 18.m06211; u6 snRNA-associated sm-like protein
Lsm2; K12621 U6 snRNA-associated Sm-like protein LSm2
Length=99
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 2 RFMNLHLKNSVCTSKDGERF---FKVAECFIRGNTIKYARLQ------EELVALVKDENP 52
+++N L N T+ D ER+ V CF+RG+ ++Y L EEL L + E
Sbjct 36 QYLNFKLSN--VTASDTERYPHLLSVVNCFVRGSVVRYVYLNSSDVNTEELQELCRREAM 93
Query 53 KAGT 56
K+
Sbjct 94 KSNV 97
> sce:YBL026W LSM2, SMX5, SNP3; Lsm (Like Sm) protein; part of
heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p):
cytoplasmic Lsm1p complex involved in mRNA decay; nuclear
Lsm8p complex part of U6 snRNP and possibly involved in processing
tRNA, snoRNA, and rRNA; K12621 U6 snRNA-associated Sm-like
protein LSm2
Length=95
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 2 RFMNLHLKNSVCTS-KDGERFFKVAECFIRGNTIKYARLQEELV 44
+F+NL L N CT K V FIRG+T++Y L + +V
Sbjct 34 QFLNLKLDNISCTDEKKYPHLGSVRNIFIRGSTVRYVYLNKNMV 77
> sce:YER112W LSM4, SDB23, USS1; Lsm (Like Sm) protein; part of
heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p):
cytoplasmic Lsm1p complex involved in mRNA decay; nuclear
Lsm8p complex part of U6 snRNP and possibly involved in processing
tRNA, snoRNA, and rRNA; K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=187
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 30/58 (51%), Gaps = 10/58 (17%)
Query 3 FMNLHLKN-------SVCTSKD---GERFFKVAECFIRGNTIKYARLQEELVALVKDE 50
+MNL L N S S+D + K+ E +IRG IK+ +LQ+ ++ VK +
Sbjct 35 WMNLTLSNVTEYSEESAINSEDNAESSKAVKLNEIYIRGTFIKFIKLQDNIIDKVKQQ 92
> cel:T10G3.6 gut-2; GUT differentiation defective family member
(gut-2); K12621 U6 snRNA-associated Sm-like protein LSm2
Length=97
Score = 31.2 bits (69), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 5/46 (10%)
Query 2 RFMNLHLKNSVCTSKDGERF---FKVAECFIRGNTIKYARLQEELV 44
+++N+ L + T D ERF V CFIRG+ ++Y +L + V
Sbjct 34 QYLNMKLTD--ITVSDPERFPHMVSVKNCFIRGSVVRYVQLPSDQV 77
> cpv:cgd5_2220 small nuclear ribonucleoprotein D1. SM domain
containing protein ; K11087 small nuclear ribonucleoprotein
D1
Length=137
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query 4 MNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELV--ALVKDENPK 53
MN +LKN V S +++ +RGNTI+Y L + L AL+ D+ PK
Sbjct 47 MNTYLKN-VKMSVKHRNPVSLSQITVRGNTIRYFILPDSLPLDALLIDDTPK 97
> ath:AT1G03330 small nuclear ribonucleoprotein D, putative /
snRNP core SM-like protein, putative / U6 snRNA-associated Sm-like
protein, putative; K12621 U6 snRNA-associated Sm-like
protein LSm2
Length=93
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 2 RFMNLHLKNSVCTSKDGE-RFFKVAECFIRGNTIKYARLQEE 42
+++N+ L+N+ +D V CFIRG+ ++Y +L ++
Sbjct 34 QYLNIKLENTRVVDQDKYPHMLSVRNCFIRGSVVRYVQLPKD 75
> tgo:TGME49_067350 small nuclear ribonucleoprotein, putative
; K11087 small nuclear ribonucleoprotein D1
Length=121
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 25/51 (49%), Gaps = 3/51 (5%)
Query 4 MNLHLKNSVCTSKDGERFFKVAECFIRGNTIKYARLQEELV--ALVKDENP 52
MN HLKN T K G + IRGN I+Y L + L L+ D+ P
Sbjct 36 MNTHLKNVKMTMKQGNP-TSLEHLTIRGNNIRYFILPDSLPLDTLLIDDTP 85
> tgo:TGME49_097140 u6 snRNA-associated sm-like protein Lsm2,
putative ; K12621 U6 snRNA-associated Sm-like protein LSm2
Length=101
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 2 RFMNLHLKN-SVCTSKDGERFFKVAECFIRGNTIKYARL 39
+F+N+ L N SV + V CFIRG+ ++Y L
Sbjct 36 QFLNIKLNNVSVADPERCPHLLSVKNCFIRGSAVRYVHL 74
> pfa:PF11_0266 small nuclear ribonucleoprotein D1, putative;
K11087 small nuclear ribonucleoprotein D1
Length=126
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 28/62 (45%), Gaps = 12/62 (19%)
Query 4 MNLHLKNSVCTSKD---------GERFFKVAECFIRGNTIKYARLQEEL---VALVKDEN 51
MN H+KN K+ ++F + IRGN I+Y L + L LV+D
Sbjct 36 MNTHMKNVKVVIKNKNIAEYNVNTKQFLSLEHVTIRGNNIRYFILSDSLPLDSLLVEDTT 95
Query 52 PK 53
PK
Sbjct 96 PK 97
> dre:57924 smx5, lsm2, wu:fe48h10, zgc:101795; smx5; K12621 U6
snRNA-associated Sm-like protein LSm2
Length=95
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query 2 RFMNLHLKN-SVCTSKDGERFFKVAECFIRGNTIKYARLQEELV--ALVKDENPKAGT 56
+++N+ L + SV + V CFIRG+ ++Y +L + V L++D K T
Sbjct 34 QYLNIKLTDISVTDPEKYPHMLSVKNCFIRGSVVRYVQLPADEVDTQLLQDAARKEAT 91
Lambda K H
0.320 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2051595972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40