bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
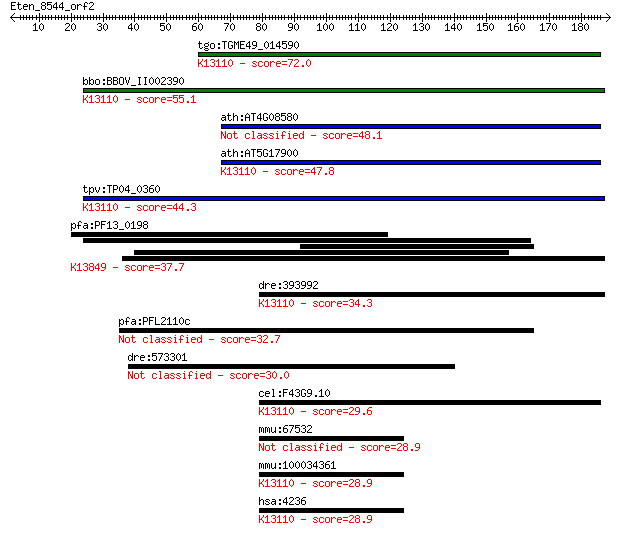
Query= Eten_8544_orf2
Length=188
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_014590 microfibrillar-associated protein 1, putativ... 72.0 1e-12
bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protei... 55.1 2e-07
ath:AT4G08580 hypothetical protein 48.1 2e-05
ath:AT5G17900 hypothetical protein; K13110 microfibrillar-asso... 47.8 2e-05
tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-asso... 44.3 2e-04
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 37.7 0.025
dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associat... 34.3 0.29
pfa:PFL2110c hypothetical protein 32.7 0.76
dre:573301 nexn, MGC103442, zgc:103442; nexilin (F actin bindi... 30.0 4.7
cel:F43G9.10 hypothetical protein; K13110 microfibrillar-assoc... 29.6 6.5
mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associa... 28.9 9.7
mmu:100034361 Mfap1b; microfibrillar-associated protein 1B; K1... 28.9 9.7
hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110 mi... 28.9 9.8
> tgo:TGME49_014590 microfibrillar-associated protein 1, putative
; K13110 microfibrillar-associated protein 1
Length=438
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 72/132 (54%), Gaps = 26/132 (19%)
Query 60 SEFSDVSEDQPEETNVVLHKPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLET 119
S ++ S+D + +LHKP FVPK R TV ERE + +E+++ ++++ RK E+
Sbjct 167 SAYTTDSDDNEDPAFTILHKPVFVPKEKRETVKERERLEAEEEKERKEAEERLADRKRES 226
Query 120 KQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSE------EMPDDTDGLDA 173
K+++Y L +EDEE + N++G + E+PDDTDGLDA
Sbjct 227 KKLLYEALQKEDEE--------------------MRTNMLGEQLQEEDCELPDDTDGLDA 266
Query 174 AQEYEDWKLREL 185
EYE WK REL
Sbjct 267 EAEYEAWKAREL 278
> bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protein
1 C-terminus containing protein; K13110 microfibrillar-associated
protein 1
Length=437
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 52/168 (30%), Positives = 91/168 (54%), Gaps = 29/168 (17%)
Query 24 DRDKLRAAVLERRRREERLLQQQQQQQQEESEESEESEFSDVSEDQPE-----ETNVVLH 78
DR ++RA + R+ EE ++Q+ + + ESE+ E +DV ++ E +T L
Sbjct 140 DRSEIRARAMAYRKEEESTIEQKTELLDQSETESED-ELTDVDTEESEPVTSSQTINALA 198
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAVLAREDEEAADAP 138
KP FVPK SRLTV E++E +R +++ + +Q+++E+R+ ++K++V L E+
Sbjct 199 KPVFVPKKSRLTVKEKKEIEREEQKKIEAEQKRLEERRKQSKELVIQTLVAEN------- 251
Query 139 VIPVEAQQQQQQQQQQQKNLMGSEEMPDDTDGLDAAQEYEDWKLRELE 186
Q+ + + N + DD D L +EYE WK+REL+
Sbjct 252 ---------MHQEIENEVNCV------DDKDEL-TEEEYELWKIRELK 283
> ath:AT4G08580 hypothetical protein
Length=435
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 62/122 (50%), Gaps = 27/122 (22%)
Query 67 EDQPEETNVVLHKPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAV 126
+D P + L KP FVPKA R T+ ERE + ++ ++++EQRK+ETKQ+V
Sbjct 173 DDMP---GIALIKPVFVPKAERDTIAERERLEAEEEALEELAKRKLEQRKIETKQIVVEE 229
Query 127 LAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSEEMPDDT---DGLDAAQEYEDWKLR 183
+ R+DEE +KN++ E D D L+ A+EYE WK R
Sbjct 230 V-RKDEEI--------------------RKNILLEEANIGDVETDDELNEAEEYEVWKTR 268
Query 184 EL 185
E+
Sbjct 269 EI 270
> ath:AT5G17900 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=435
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 62/122 (50%), Gaps = 27/122 (22%)
Query 67 EDQPEETNVVLHKPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAV 126
+D P + + KP FVPKA R T+ ERE + ++ ++++EQRKLETKQ+V
Sbjct 173 DDMP---GIAMIKPVFVPKAERDTIAERERLEAEEEALEELAKRKLEQRKLETKQIVVEE 229
Query 127 LAREDEEAADAPVIPVEAQQQQQQQQQQQKNLMGSEEMPDDT---DGLDAAQEYEDWKLR 183
+ R+DEE +KN++ E D D L+ A+EYE WK R
Sbjct 230 V-RKDEEI--------------------RKNILLEEANIGDVETDDELNEAEEYEVWKTR 268
Query 184 EL 185
E+
Sbjct 269 EI 270
> tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=408
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 81/163 (49%), Gaps = 25/163 (15%)
Query 24 DRDKLRAAVLERRRREERLLQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHKPAFV 83
DR LR LE R+REE + +++ EESE E ++ E VL KP FV
Sbjct 126 DRSTLRKLALEYRKREEESAPKTVEEEVEESESDSSEESEYQEDEAGPEDLDVLSKPVFV 185
Query 84 PKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVE 143
PK SR T E+E+ ++ ++ +++++ +RK +TK+MV
Sbjct 186 PKGSRKTESEKEQLRKEEVLRRENEKKRLMERKKDTKEMVI------------------- 226
Query 144 AQQQQQQQQQQQKNLMGSEEMPDDTDGLDAAQEYEDWKLRELE 186
Q+ Q+ ++ +E+ DDTD D +EYE WK+REL+
Sbjct 227 -----QKVQELEEEPEPEDELIDDTDTFD-EKEYELWKIRELK 263
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 37.7 bits (86), Expect = 0.025, Method: Composition-based stats.
Identities = 26/101 (25%), Positives = 56/101 (55%), Gaps = 2/101 (1%)
Query 20 ETITDRDKLRAAVLERRRREERLLQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHK 79
E + ++L+ ER ++EE L +Q+Q++ Q+E E + + E Q + K
Sbjct 2767 EQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELK 2826
Query 80 PAFVPKASRLTVMEREEQQRRVREEQ--QQQQQQVEQRKLE 118
+ + ++R+EQ+R +EE+ +Q+Q+++E++K+E
Sbjct 2827 RQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERKKIE 2867
Score = 36.2 bits (82), Expect = 0.074, Method: Composition-based stats.
Identities = 34/142 (23%), Positives = 75/142 (52%), Gaps = 14/142 (9%)
Query 24 DRDKLRAAVLERRRREERLLQQQQQQ-QQEESEESEESEFSDVSEDQPEETNVVLHKPAF 82
++++L+ +R+ +ERL +++Q+Q Q+EE + +E E E + L K
Sbjct 2743 EQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKE-- 2800
Query 83 VPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVY-AVLAREDEEAADAPVIP 141
++R+EQ+R RE+Q+Q Q++ E ++ E +++ L R+++E
Sbjct 2801 -------EELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQK---E 2850
Query 142 VEAQQQQQQQQQQQKNLMGSEE 163
E ++Q+Q++ +++K + E
Sbjct 2851 EELKRQEQERLERKKIELAERE 2872
Score = 34.7 bits (78), Expect = 0.21, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 92 MEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQ 151
++R+EQ+R RE+Q+Q Q++ E ++ E +++ + E+ ++ Q+Q++ +
Sbjct 2753 LKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLE 2812
Query 152 QQQQKNLMGSEEM 164
+++Q+ L EE+
Sbjct 2813 REKQEQLQKEEEL 2825
Score = 30.4 bits (67), Expect = 3.3, Method: Composition-based stats.
Identities = 25/119 (21%), Positives = 61/119 (51%), Gaps = 5/119 (4%)
Query 40 ERLLQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHKPAFVPKASRLTVMEREEQQR 99
+R Q++ Q+++E + +E + E +E + + + K L +R+EQ+R
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEAL---KRQEQER 2796
Query 100 RVREEQ--QQQQQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQQQK 156
+EE+ +Q+Q+++E+ K E Q + +E E + + Q++ Q++++ ++
Sbjct 2797 LQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKR 2855
Score = 29.6 bits (65), Expect = 6.1, Method: Composition-based stats.
Identities = 32/153 (20%), Positives = 76/153 (49%), Gaps = 27/153 (17%)
Query 36 RRREERLLQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHKPAFVPKASRLTVMERE 95
+R+E+ LQ++++ +++E E E + + +++ ++R+
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEE---------------------LKRQ 2778
Query 96 EQQRRVREE--QQQQQQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQQQQ 153
EQ+R +EE ++Q+Q+++ Q++ E K+ L RE +E E ++Q+Q++ Q
Sbjct 2779 EQERLQKEEALKRQEQERL-QKEEELKRQEQERLEREKQEQLQK---EEELKRQEQERLQ 2834
Query 154 QQKNLMGSEEMPDDTDGLDAAQEYEDWKLRELE 186
+++ L E+ + QE E + +++E
Sbjct 2835 KEEALKRQEQERLQKEEELKRQEQERLERKKIE 2867
> dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associated
protein 1; K13110 microfibrillar-associated protein 1
Length=437
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 59/108 (54%), Gaps = 22/108 (20%)
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAVLAREDEEAADAP 138
KP F+ K R+TV ERE +++R +E + + ++Q E+R+ T ++V
Sbjct 199 KPVFIRKKDRVTVAEREAEEQRQKELEVEAKKQAEERRRYTLKIV--------------- 243
Query 139 VIPVEAQQQQQQQQQQQKNLMGSEEMPDDTDGLDAAQEYEDWKLRELE 186
++ +++ ++ ++ L E + DTDG + +EYE WK+REL+
Sbjct 244 -----EEEAKKEFEENKRTLAALEAL--DTDGENEEEEYEAWKVRELK 284
> pfa:PFL2110c hypothetical protein
Length=1846
Score = 32.7 bits (73), Expect = 0.76, Method: Composition-based stats.
Identities = 27/134 (20%), Positives = 69/134 (51%), Gaps = 16/134 (11%)
Query 35 RRRREERL-LQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHKPAFVPKASRLTVME 93
RR EE++ + +++Q+ EE + E V E+Q V + +L +
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQK------------VGEEQKLREEQ 1534
Query 94 REEQQRRVREEQ---QQQQQQVEQRKLETKQMVYAVLAREDEEAADAPVIPVEAQQQQQQ 150
+ +++++REEQ ++Q+ + EQ+ E ++M RE+++ + + E + +++Q
Sbjct 1535 KMREEQKMREEQKMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQ 1594
Query 151 QQQQQKNLMGSEEM 164
+ ++++ + ++M
Sbjct 1595 KMREEQKMREEQKM 1608
> dre:573301 nexn, MGC103442, zgc:103442; nexilin (F actin binding
protein)
Length=861
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 57/116 (49%), Gaps = 16/116 (13%)
Query 38 REERLLQQQQQQQQEESEESEESEFSDVSEDQPEETNVVLHKP---AFVPKASRLTVMER 94
+EE Q + +++ +ES + + + SDV+ Q E + KP +++PK + V +
Sbjct 90 KEEDTSQLENEEKADESHDIDITNMSDVALKQ--EKLLKTSKPVQRSYIPKLGKEDVKNK 147
Query 95 EEQQRRVREEQQQQQQQVEQRKL-----------ETKQMVYAVLAREDEEAADAPV 139
E ++ REE+ Q++ Q EQ+K KQM+ +LA DEE P
Sbjct 148 FEAMQKAREERNQKRSQEEQKKRREQYVKEREYGRRKQMIKELLASSDEEEEVKPT 203
> cel:F43G9.10 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=466
Score = 29.6 bits (65), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 58/107 (54%), Gaps = 19/107 (17%)
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMVYAVLAREDEEAADAP 138
KP F K R+T+ E E+++ + ++ + +++ E+RK E+ ++V VL ++EEAA
Sbjct 225 KPIFTRKKDRITLQEAEKEKEKEILKKIEDEKRAEERKRESAKLVEKVL--QEEEAA--- 279
Query 139 VIPVEAQQQQQQQQQQQKNLMGSEEMPDDTDGLDAAQEYEDWKLREL 185
++++ + + + S D+T+ + YE WKLRE+
Sbjct 280 ----------EKRKTEDRVDLSSVLTDDETENM----AYEAWKLREM 312
> mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associated
protein 1A
Length=439
Score = 28.9 bits (63), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMV 123
KP F+ K R+TV ERE + + +E +Q+ ++ E+R+ T ++V
Sbjct 200 KPVFIRKKDRVTVQEREAEALKQKELEQEAKRMAEERRKYTLKIV 244
> mmu:100034361 Mfap1b; microfibrillar-associated protein 1B;
K13110 microfibrillar-associated protein 1
Length=439
Score = 28.9 bits (63), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMV 123
KP F+ K R+TV ERE + + +E +Q+ ++ E+R+ T ++V
Sbjct 200 KPVFIRKKDRVTVQEREAEALKQKELEQEAKRMAEERRKYTLKIV 244
> hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110
microfibrillar-associated protein 1
Length=439
Score = 28.9 bits (63), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 79 KPAFVPKASRLTVMEREEQQRRVREEQQQQQQQVEQRKLETKQMV 123
KP F+ K R+TV ERE + + +E +Q+ ++ E+R+ T ++V
Sbjct 200 KPVFIRKKDRVTVQEREAEALKQKELEQEAKRMAEERRKYTLKIV 244
Lambda K H
0.306 0.120 0.304
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5300054584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40