bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8567_orf2
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
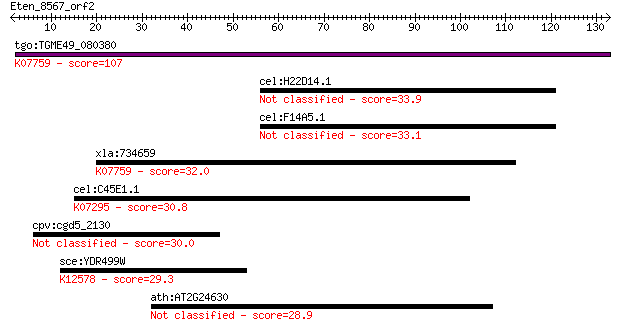
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 107 1e-23
cel:H22D14.1 nhr-267; Nuclear Hormone Receptor family member (... 33.9 0.13
cel:F14A5.1 nhr-264; Nuclear Hormone Receptor family member (n... 33.1 0.20
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 32.0 0.45
cel:C45E1.1 nhr-64; Nuclear Hormone Receptor family member (nh... 30.8 1.2
cpv:cgd5_2130 chromatin associated proein with a chromodomain ... 30.0 1.9
sce:YDR499W LCD1, DDC2, PIE1; Essential protein required for t... 29.3 3.4
ath:AT2G24630 ATCSLC08; cellulose synthase/ transferase, trans... 28.9 4.9
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 55/148 (37%), Positives = 84/148 (56%), Gaps = 17/148 (11%)
Query 2 GNWRVDKPLRI---GPLTHHLATHKNLSKSLLPFLASLARRLPDFFPSGIRHLGRDNPAV 58
GN R +K RI G L + LPF+A+L R+ + FPSG++++ +NP V
Sbjct 91 GNIRTEKNKRILQRGLLNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPENPQV 150
Query 59 HLRRIEVLALLAALFFGIV--------------YPLQMNAPDMHYRGFFERPPKYQSLLQ 104
HLR+I+V L+AA F G++ + + N DM YRGF ER PK+QS+L
Sbjct 151 HLRKIQVFTLIAAAFLGVIPHNQRALLAHHQKKFVMNQNKLDMFYRGFMERKPKFQSVLI 210
Query 105 YFQQMQQTVGSCWQQMLQRGDVGPLRCS 132
YF M++ +G+CW +M+++G V C+
Sbjct 211 YFASMRERLGNCWNEMVKKGFVDMAPCT 238
> cel:H22D14.1 nhr-267; Nuclear Hormone Receptor family member
(nhr-267)
Length=344
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 56 PAVH-LRRIEVLALLAALFFGIVYPLQMNAPDMHYRGFFERPPKYQSLLQYFQQMQQTVG 114
P V+ L + +V LL F V+ + A M +E PK + L +Y QQ++ T+G
Sbjct 204 PGVNSLEKEDVQTLLKYFQFANVWMDSVRAYSMSNVDIYETTPKDKRLSEYIQQVKLTLG 263
Query 115 SCWQQM 120
S + Q+
Sbjct 264 SSFSQL 269
> cel:F14A5.1 nhr-264; Nuclear Hormone Receptor family member
(nhr-264)
Length=408
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 56 PAVH-LRRIEVLALLAALFFGIVYPLQMNAPDMHYRGFFERPPKYQSLLQYFQQMQQTVG 114
P V+ L + +V LL F V+ + A M +E PK + L +Y QQ++ T+G
Sbjct 254 PGVNSLEKEDVQTLLKYFQFANVWMDSVRAYSMSNVDIYETTPKDKRLSEYIQQVKLTLG 313
Query 115 SCWQQM 120
S + Q+
Sbjct 314 SSFSQL 319
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 27/107 (25%), Positives = 54/107 (50%), Gaps = 17/107 (15%)
Query 20 ATHKNLSKSLLPFLASLARRLPDFFPSGIRHLGRD-NPAVHLRRIEVLALLAALFFGIVY 78
A + +L S+LP + +LA LP+ L + N ++ + ++++ +LLA FF +
Sbjct 381 AEYDHLFHSILPDMVNLALSLPNICTQPTPLLKQKMNHSITMSQMQIASLLANAFF-CTF 439
Query 79 PLQMNA---------PDMHYRGFFE-----RPPKYQSLLQYFQQMQQ 111
P + NA PD+++ FE + K ++L YF+++ +
Sbjct 440 P-RRNARMKSEYSSYPDINFNRLFEGKNPKKAEKLKTLFCYFRRVTE 485
> cel:C45E1.1 nhr-64; Nuclear Hormone Receptor family member (nhr-64);
K07295 nuclear receptor subfamily 2 group A
Length=369
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 41/94 (43%), Gaps = 16/94 (17%)
Query 15 LTHHLATHKNLSKSLLPFLASLARRLPDFFPSGIRHLGRDNPAVHLRRIEVLALLAALFF 74
LT+ HK+ K +P + +A R+ D + +R L H+ IE +AL A FF
Sbjct 216 LTNETCLHKDSPK--IPDMNRVAERIIDQVTNPMRSL-------HMNEIEYIALKAIAFF 266
Query 75 -----GIVYPLQMNAPDMHYRGF--FERPPKYQS 101
GI + +M R FER +Y S
Sbjct 267 DPLAKGITSESYSDVEEMRQRILESFERHVRYVS 300
> cpv:cgd5_2130 chromatin associated proein with a chromodomain
at the C-terminus
Length=1075
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 6 VDKPLRIGPLTHHLATHKNLSKSLLPFLASLARRLPDFFPS 46
+ P+ + P THH+ K LS S L S+ R P + S
Sbjct 494 IQNPIILAPKTHHMIVKKALSNSRYWSLDSITSRCPQSYTS 534
> sce:YDR499W LCD1, DDC2, PIE1; Essential protein required for
the DNA integrity checkpoint pathways; interacts physically
with Mec1p; putative homolog of S. pombe Rad26 and human ATRIP;
K12578 DNA damage checkpoint protein LCD1
Length=747
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 12 IGPLTHHLATHKNLSKSLLPFLASLARRLPDFFPSGIRHLG 52
I L ++ H N SK +PFL +L ++ F PS +L
Sbjct 278 IAVLIKEISVHPNESKLAVPFLVALMYQIVQFRPSATHNLA 318
> ath:AT2G24630 ATCSLC08; cellulose synthase/ transferase, transferring
glycosyl groups
Length=690
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 37/85 (43%), Gaps = 19/85 (22%)
Query 32 FLASLARRLPDFFPSGIRHLGRDNPAVHLRRIEVLALLAALFFGIVYPLQMNAPDMHYRG 91
L S+ RRL P G LGRD ++ ++A LA L F +V +YRG
Sbjct 81 LLGSVKRRLSFTHPLGSERLGRDGWLFSAIKLFLVASLAILAFELV---------AYYRG 131
Query 92 --FFERPP--------KYQSLLQYF 106
+F+ P + QSLL F
Sbjct 132 WHYFKNPNLHIPTSKLEIQSLLHLF 156
Lambda K H
0.328 0.143 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40