bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8597_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
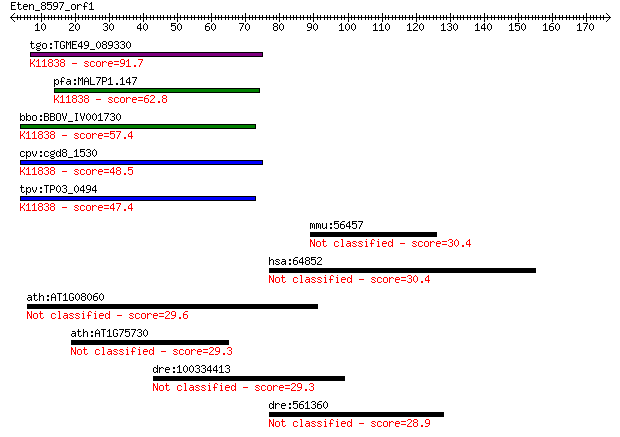
tgo:TGME49_089330 ubiquitin carboxyl-terminal hydrolase, putat... 91.7 1e-18
pfa:MAL7P1.147 ubiquitin carboxyl-terminal hydrolase, putative... 62.8 6e-10
bbo:BBOV_IV001730 21.m03027; ubiquitin carboxyl-terminal hydro... 57.4 2e-08
cpv:cgd8_1530 ubiquitin carboxyl terminal hydrolase domain tha... 48.5 1e-05
tpv:TP03_0494 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.... 47.4 2e-05
mmu:56457 Clptm1, HS9, N14; cleft lip and palate associated tr... 30.4 3.2
hsa:64852 TUT1, FLJ21850, FLJ22267, FLJ22347, MGC131987, MGC14... 30.4 3.5
ath:AT1G08060 MOM; MOM (MORPHEUS MOLECULE) 29.6 5.8
ath:AT1G75730 hypothetical protein 29.3 6.9
dre:100334413 enzymatic polyprotein; Endonuclease; Reverse tra... 29.3 7.5
dre:561360 hypothetical LOC561360 28.9 9.7
> tgo:TGME49_089330 ubiquitin carboxyl-terminal hydrolase, putative
(EC:1.1.3.12 3.1.2.15); K11838 ubiquitin carboxyl-terminal
hydrolase 7 [EC:3.1.2.15]
Length=2100
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/77 (61%), Positives = 54/77 (70%), Gaps = 9/77 (11%)
Query 7 NYLRGE---------RKPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILSRCRT 57
NYL GE R PR+R K YNAYILVYV K LA LLADC+P+KVNPQ++ RCRT
Sbjct 819 NYLAGEFRFFRLQSSRAPRSRQKVYNAYILVYVKKRLANQLLADCNPMKVNPQVVIRCRT 878
Query 58 EEQLRRLRTSIRNVLEK 74
EE L +LR IR VLE+
Sbjct 879 EELLMKLRNRIRRVLEQ 895
> pfa:MAL7P1.147 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15); K11838 ubiquitin carboxyl-terminal hydrolase
7 [EC:3.1.2.15]
Length=3183
Score = 62.8 bits (151), Expect = 6e-10, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 14 KPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILSRCRTEEQLRRLRTSIRN-VL 72
K + R K Y+AY+LVYV K+L P L+ +C P VNPQ++ RCR EE + + R +++ +L
Sbjct 1181 KIKQRLKNYSAYMLVYVKKSLIPKLIKECDPAVVNPQVVKRCRLEEIINKKRYKLKDEIL 1240
Query 73 E 73
E
Sbjct 1241 E 1241
> bbo:BBOV_IV001730 21.m03027; ubiquitin carboxyl-terminal hydrolase;
K11838 ubiquitin carboxyl-terminal hydrolase 7 [EC:3.1.2.15]
Length=1446
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 4 ECYNYLRGER----KPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILSRCRTEE 59
+CYNY E K R K YNAYIL+YV K +L C P+K + +++RC EE
Sbjct 561 DCYNYCESEDNASVKAFRRPKNYNAYILIYVLKDAVDDILGPCDPVKEHYSMITRCSLEE 620
Query 60 QLRRLRTSIRNVL 72
+L LR +R L
Sbjct 621 RLYNLRHRVRERL 633
> cpv:cgd8_1530 ubiquitin carboxyl terminal hydrolase domain that
is fused to a MATH domain ; K11838 ubiquitin carboxyl-terminal
hydrolase 7 [EC:3.1.2.15]
Length=1607
Score = 48.5 bits (114), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query 4 ECYNYL-RGERKPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILSRCRTEEQLR 62
E ++YL E R+K ++AYILVYV + A LL++ P +VNP ++S+ R E ++
Sbjct 648 EVWDYLGNPESDIPKRSKTHSAYILVYVREDQAQDLLSEPIPQEVNPDLVSKYRKEIEMM 707
Query 63 RLRTSIRNVLEK 74
LR +R L +
Sbjct 708 NLRKRLRQDLNE 719
> tpv:TP03_0494 ubiquitin carboxyl-terminal hydrolase (EC:3.1.2.15);
K11838 ubiquitin carboxyl-terminal hydrolase 7 [EC:3.1.2.15]
Length=1062
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 38/79 (48%), Gaps = 10/79 (12%)
Query 4 ECYNYL----------RGERKPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILS 53
+CYNY R+K YNAYIL+YV K+ +L + LK N Q+LS
Sbjct 654 DCYNYFTTNPSNPSIYSNHMNRYRRSKVYNAYILIYVLKSKKDEILGEVDMLKENYQMLS 713
Query 54 RCRTEEQLRRLRTSIRNVL 72
R R + L LR+ R L
Sbjct 714 RYRIQRLLHCLRSKTRERL 732
> mmu:56457 Clptm1, HS9, N14; cleft lip and palate associated
transmembrane protein 1
Length=664
Score = 30.4 bits (67), Expect = 3.2, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 89 PLPGTAVPVCDSDTGPKAQSGQPAAAAARDDKPRAHE 125
P P TAV D+ T PKA SG A+ ++ P+ E
Sbjct 623 PAPSTAVSGEDASTVPKATSGACTASQPQEAPPKPAE 659
> hsa:64852 TUT1, FLJ21850, FLJ22267, FLJ22347, MGC131987, MGC149809,
PAPD2, RBM21, STARPAP, TUTase; terminal uridylyl transferase
1, U6 snRNA-specific (EC:2.7.7.19 2.7.7.52)
Length=912
Score = 30.4 bits (67), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 33/78 (42%), Gaps = 1/78 (1%)
Query 77 GQGLPPAAVQVPPLPGTAVPVCDSDTGPKAQSGQPAAAAARDDKPRAHEAALRRRGQLEG 136
G+ + A+Q P PG +P+ G + GQP+ AA + P+ HEAA G
Sbjct 735 GEMVQDWAMQSPGQPGD-LPLTTGKHGAPGEEGQPSHAALAERGPKGHEAAQEWSQGEAG 793
Query 137 VPGVLRVSVSGFSGEWRR 154
L S S W R
Sbjct 794 KGASLPSSASWRCALWHR 811
> ath:AT1G08060 MOM; MOM (MORPHEUS MOLECULE)
Length=2001
Score = 29.6 bits (65), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 43/89 (48%), Gaps = 5/89 (5%)
Query 6 YNYLRGERKPRTRNKPYNAYILVYVNKTLAPSLLADCSPLKVNPQILSRCRTEEQLRRLR 65
++ + E RT K LV +NK LA + L+ C+ KV+P R + ++ +R
Sbjct 1780 FHEVEAEHNTRT-TKIEKDKNLVIMNKLLANAFLSKCTDKKVSPSGAPRGKIQQLAQRAA 1838
Query 66 --TSIRNVL--EKCQGQGLPPAAVQVPPL 90
+++RN + ++ Q P A+ PL
Sbjct 1839 QVSALRNYIAPQQLQASSFPAPALVSAPL 1867
> ath:AT1G75730 hypothetical protein
Length=589
Score = 29.3 bits (64), Expect = 6.9, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 19 NKPYNAYILVYVNKTLAPS---LLADCSPLKVNPQILSRCRTEEQLRRL 64
+KP + + ++ + ++PS L +C PL+V P+ L RC + + RL
Sbjct 281 SKPSSTKLPPWMGQAVSPSNTASLLNCEPLRVQPRKLKRCASHIYISRL 329
> dre:100334413 enzymatic polyprotein; Endonuclease; Reverse transcriptase,
putative-like
Length=1766
Score = 29.3 bits (64), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 43 SPLKVNPQILSRCRTEEQLRRLRTSIRNVLEKCQGQGLPPAAVQVPPLPGTAVPVC 98
SP +V Q L +EE L+RL ++ G G+ P Q PP P T VP C
Sbjct 997 SPTRVIKQ-LKENHSEEWLQRLARYTTQCVDFLSGPGVLPITFQEPPAP-TVVPSC 1050
> dre:561360 hypothetical LOC561360
Length=1047
Score = 28.9 bits (63), Expect = 9.7, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 22/51 (43%), Gaps = 0/51 (0%)
Query 77 GQGLPPAAVQVPPLPGTAVPVCDSDTGPKAQSGQPAAAAARDDKPRAHEAA 127
G+ P Q+P P VP +S T P Q+ Q A D+P +A
Sbjct 967 GKITPVRPTQIPQRPTWTVPTPNSPTSPTNQNQQTAPRIKPPDQPNVSRSA 1017
Lambda K H
0.317 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40