bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8635_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
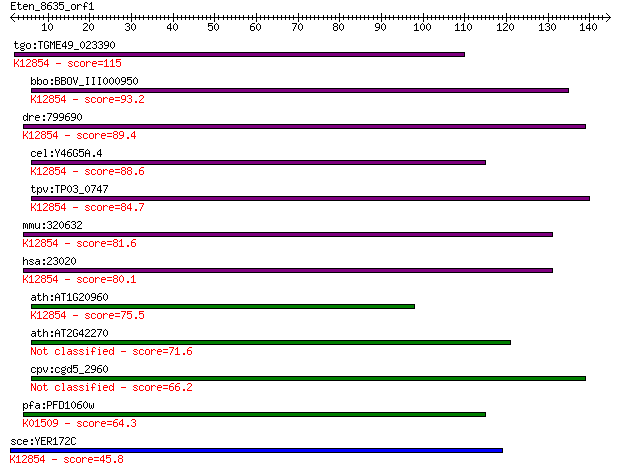
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 115 5e-26
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 93.2 2e-19
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 89.4 3e-18
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 88.6 6e-18
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 84.7 8e-17
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 81.6 7e-16
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 80.1 2e-15
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 75.5 5e-14
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 71.6 7e-13
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 66.2 3e-11
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 64.3 1e-10
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 45.8 4e-05
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 115 bits (288), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 68/110 (61%), Positives = 85/110 (77%), Gaps = 2/110 (1%)
Query 2 ENFFYRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGI 61
E FFY+PTT+ T VYEQLLV LQQQLGDQPDEVL+GAADEVL LK DG +E E+K+ +
Sbjct 97 EIFFYKPTTQQTRLVYEQLLVTLQQQLGDQPDEVLKGAADEVLAALKADGCRESEKKKNV 156
Query 62 EAVLGPLTQERFTKLFQLSKNITDFM--GADQEGDEAGEPDATAGVAVVF 109
E VLGP+ + FT+L QL+KNITD+ G +++G +AG D + GVAVVF
Sbjct 157 EQVLGPVNSDVFTRLHQLAKNITDYSLGGEEEDGAQAGGLDGSTGVAVVF 206
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 52/129 (40%), Positives = 79/129 (61%), Gaps = 1/129 (0%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y P++ +T YE++L +LQ+ L DQP +VLR A EV+ L+ GLK +R++ E +L
Sbjct 102 YTPSSVSTRVKYEEILGILQECLTDQPHDVLRDAFGEVMTHLRSPGLKSEDRRKLCEEIL 161
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEEGEEDYDYSDME 125
GP+T ++F +L+ SK++ DF +D D G+AVVFDE+E+ E D + D E
Sbjct 162 GPITDDQFYRLYHASKDLVDFGDSDAAPGTNEFTDNPMGIAVVFDEDEDAESDVNSVD-E 220
Query 126 AEGDEEEDE 134
DE+ DE
Sbjct 221 DYLDEDHDE 229
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 56/136 (41%), Positives = 81/136 (59%), Gaps = 1/136 (0%)
Query 4 FFYRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEA 63
Y+P T+ T + YE LL +Q LGDQP ++L GAADEVL VLK D +++ ER+R +E
Sbjct 100 IVYKPKTKETRETYEILLSFIQAALGDQPRDILCGAADEVLAVLKNDKMRDKERRREVEQ 159
Query 64 VLGPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEEGEEDYDYSD 123
+LGP R+ L L K I+D+ G + + D T GV V F+ +EE ++ Y +
Sbjct 160 LLGPTDDTRYHVLVNLGKKISDYGGDKELQNMDDNIDETYGVNVQFESDEEEGDEDQYGE 219
Query 124 MEAEG-DEEEDEETEE 138
+ EG D+ E EE +E
Sbjct 220 VRDEGSDDSEGEEADE 235
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 88.6 bits (218), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 52/111 (46%), Positives = 68/111 (61%), Gaps = 3/111 (2%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y+P T+ T Q YE +L + LGD P EVL GAADEVL LK D ++ E+K+ +EA+L
Sbjct 97 YKPRTQETKQTYEVILSFILDALGDVPREVLCGAADEVLLTLKNDKFRDKEKKKEVEALL 156
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDA--TAGVAVVFDEEEE 114
GPLT +R L LSK I+DF ++E G+ D GV V FD +EE
Sbjct 157 GPLTDDRIAVLINLSKKISDF-SIEEENKPEGDGDIYENEGVNVQFDSDEE 206
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 84.7 bits (208), Expect = 8e-17, Method: Composition-based stats.
Identities = 51/134 (38%), Positives = 80/134 (59%), Gaps = 5/134 (3%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y P + T Y+++L +LQ+ LGDQP VL A DE L LK + E ER+ +E VL
Sbjct 101 YVPRSGYTRTKYQEMLTILQKILGDQPQSVLLSACDETLLSLKTENSAE-ERRALVEDVL 159
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEEGEEDYDYSDME 125
GP++ E + LF LSK +TDF A ++ D G+ G+AV+FDE++ + + ++
Sbjct 160 GPVSDEVYYNLFHLSKELTDFKMASEQ-DTGGDTFDDTGLAVIFDEDDNQDTN---EVVD 215
Query 126 AEGDEEEDEETEEQ 139
EG+E+++ E + Q
Sbjct 216 PEGEEDDENELDPQ 229
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 81.6 bits (200), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 56/134 (41%), Positives = 76/134 (56%), Gaps = 7/134 (5%)
Query 4 FFYRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEA 63
Y+P T+ T + YE LL +Q LGDQP ++L GAADEVL VLK + L++ ER+R I+
Sbjct 100 IIYKPKTKETRETYEVLLSFIQAALGDQPRDILCGAADEVLAVLKNEKLRDKERRREIDL 159
Query 64 VLGPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEE-------GE 116
+LG R+ L L K ITD+ G + + D T GV V F+ +EE GE
Sbjct 160 LLGQTDDTRYHVLVNLGKKITDYGGDKEIQNMDDNIDETYGVNVQFESDEEEGDEDVYGE 219
Query 117 EDYDYSDMEAEGDE 130
+ SD + EGDE
Sbjct 220 VREEASDDDMEGDE 233
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 76/134 (56%), Gaps = 7/134 (5%)
Query 4 FFYRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEA 63
Y+P T+ T + YE LL +Q LGDQP ++L GAADEVL VLK + L++ ER++ I+
Sbjct 100 IIYKPKTKETRETYEVLLSFIQAALGDQPRDILCGAADEVLAVLKNEKLRDKERRKEIDL 159
Query 64 VLGPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEE-------GE 116
+LG R+ L L K ITD+ G + + D T GV V F+ +EE GE
Sbjct 160 LLGQTDDTRYHVLVNLGKKITDYGGDKEIQNMDDNIDETYGVNVQFESDEEEGDEDVYGE 219
Query 117 EDYDYSDMEAEGDE 130
+ SD + EGDE
Sbjct 220 VREEASDDDMEGDE 233
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 40/92 (43%), Positives = 56/92 (60%), Gaps = 4/92 (4%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y+P T+ T YE +L L+Q+QLG QP ++ GAADE+L VLK D + E+K IE +L
Sbjct 110 YQPKTKETRAAYEAMLGLIQKQLGGQPPSIVSGAADEILAVLKNDAFRNPEKKMEIEKLL 169
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEAG 97
+ F +L + K ITDF QEG ++G
Sbjct 170 NKIENHEFDQLVSIGKLITDF----QEGGDSG 197
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 45/115 (39%), Positives = 68/115 (59%), Gaps = 14/115 (12%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y+P T+ T +E +L L+QQQLG QP +++ GAADE+L VLK + +K E+K IE +L
Sbjct 110 YQPKTKETRVAFEIMLGLIQQQLGGQPLDIVCGAADEILAVLKNESVKNHEKKVEIEKLL 169
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEEGEEDYD 120
+T + F++ + K ITD+ +E G D+ +G A E+G DYD
Sbjct 170 NVITDQVFSQFVSIGKLITDY-------EEGG--DSLSGKA-----SEDGGLDYD 210
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 43/135 (31%), Positives = 72/135 (53%), Gaps = 4/135 (2%)
Query 6 YRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEAVL 65
Y PTT T Y++LL +L LG QP ++L A E+L VLK D L++ E+ + + ++
Sbjct 94 YTPTTMETRLAYDELLGILTDILGSQPSKILEDTAFELLIVLKNDELRDEEKYKKCKEII 153
Query 66 GPLTQERFTKLFQLSKNITDFMGADQEGDEA--GEPDATAGVAVVFDEEEEGEEDYDYSD 123
L ++ + S+ I DF +E D++ + + + VAV+FDE + + + SD
Sbjct 154 IQLNNTTYSDILDKSRRIVDFTLNMEEMDDSQNKQTEEASEVAVIFDESDSEKRKGELSD 213
Query 124 MEAEGDEEEDEETEE 138
+ D E D +T E
Sbjct 214 --GDDDSENDSDTSE 226
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 39/112 (34%), Positives = 67/112 (59%), Gaps = 5/112 (4%)
Query 4 FFYRPTTRATNQVYEQLLVLLQQQLGDQPDEVLRGAADEVLQVLKEDGLKEGERKRGIEA 63
F Y+PTT+ T ++Y +L+ ++ LGD +++ A +E+L +LK + L E+K+ +E+
Sbjct 111 FLYKPTTKYTEEIYTKLMSKIRFLLGDNTGDIINSACNEILYILKNEELNNEEKKKQVES 170
Query 64 VLGP-LTQERFTKLFQLSKNITDFMGADQEGDEAGEPDATAGVAVVFDEEEE 114
L + + F ++ LSK I DF QE E E D GVAV+F+E+++
Sbjct 171 ELEIYINDDIFIEINNLSKEIYDF--NKQEEGEYVEND--EGVAVIFEEDDD 218
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 40/126 (31%), Positives = 63/126 (50%), Gaps = 17/126 (13%)
Query 1 DENFFYRPTTRATNQVYEQLLVLLQQQLG-DQPDEVLRGAADEVLQVLKEDGLKEG---- 55
D Y P + + YEQ+L + + LG D P +++ G AD ++ LKE+ E
Sbjct 104 DFRLHYYPKDPSNVETYEQILQWVTEVLGNDIPHDLIIGTADIFIRQLKENEENEDGNIE 163
Query 56 ERKRGIEAVLG-PLTQERFTKLFQLSKNITDFMGADQEGDEAGEPDAT--AGVAVVFDEE 112
ERK I+ LG + +F +L +L KNITD+ PD + VA++ D+E
Sbjct 164 ERKEKIQHELGINIDSLKFNELVKLMKNITDY---------ETHPDNSNKQAVAILADDE 214
Query 113 EEGEED 118
+ EE+
Sbjct 215 KSDEEE 220
Lambda K H
0.306 0.131 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40