bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8674_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
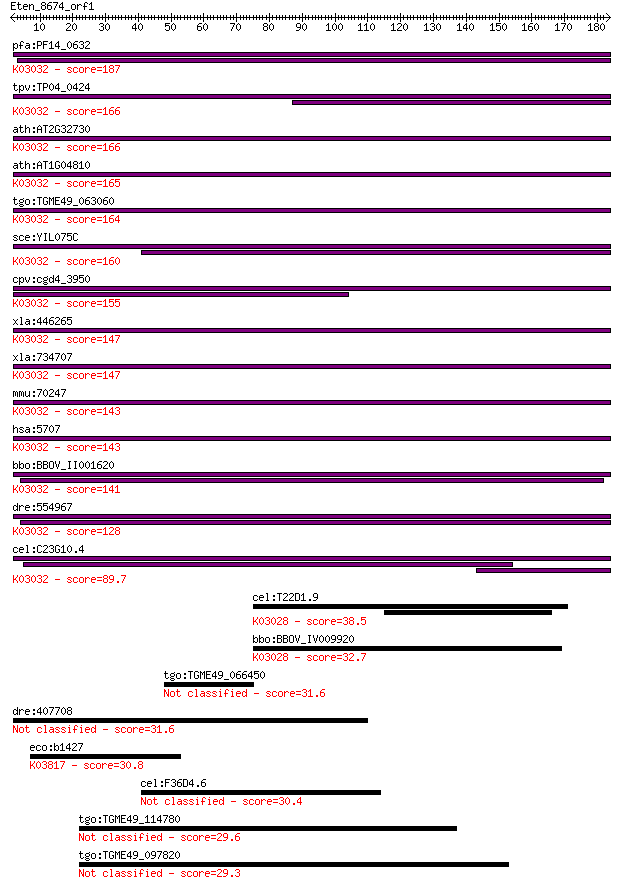
pfa:PF14_0632 26S proteasome subunit, putative; K03032 26S pro... 187 1e-47
tpv:TP04_0424 26S proteasome regulatory subunit; K03032 26S pr... 166 3e-41
ath:AT2G32730 26S proteasome regulatory subunit, putative; K03... 166 6e-41
ath:AT1G04810 26S proteasome regulatory subunit, putative; K03... 165 9e-41
tgo:TGME49_063060 26S proteasome non-ATPase regulatory subunit... 164 1e-40
sce:YIL075C RPN2, SEN3; Rpn2p; K03032 26S proteasome regulator... 160 2e-39
cpv:cgd4_3950 RPN2/26s proteasome regulatory subunit ; K03032 ... 155 9e-38
xla:446265 psmd1; proteasome (prosome, macropain) 26S subunit,... 147 2e-35
xla:734707 hypothetical protein MGC114631; K03032 26S proteaso... 147 3e-35
mmu:70247 Psmd1, 2410026J11Rik, P112, S1; proteasome (prosome,... 143 4e-34
hsa:5707 PSMD1, MGC133040, MGC133041, P112, Rpn2, S1; proteaso... 143 4e-34
bbo:BBOV_II001620 18.m09940; rpn2_yeast 26S proteasome regulat... 141 1e-33
dre:554967 psmd1, MGC55818, zgc:55818; proteasome (prosome, ma... 128 8e-30
cel:C23G10.4 rpn-2; proteasome Regulatory Particle, Non-ATPase... 89.7 5e-18
cel:T22D1.9 rpn-1; proteasome Regulatory Particle, Non-ATPase-... 38.5 0.013
bbo:BBOV_IV009920 23.m05991; proteasome 26S regulatory subunit... 32.7 0.77
tgo:TGME49_066450 lysine decarboxylase domain-containing protein 31.6 1.6
dre:407708 lmf2b, im:7150505, im:7152257, lmf2, tmem112b, tmem... 31.6 1.7
eco:b1427 rimL, ECK1420, JW1423; ribosomal-protein-L7/L12-seri... 30.8 3.0
cel:F36D4.6 hypothetical protein 30.4 3.8
tgo:TGME49_114780 myosin G 29.6 5.4
tgo:TGME49_097820 armadillo/beta-catenin-like repeat protein 29.3 8.8
> pfa:PF14_0632 26S proteasome subunit, putative; K03032 26S proteasome
regulatory subunit N2
Length=1172
Score = 187 bits (476), Expect = 1e-47, Method: Composition-based stats.
Identities = 96/186 (51%), Positives = 131/186 (70%), Gaps = 7/186 (3%)
Query 2 LYGLGLIHCG--APNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC 59
LY LGLIH + +++ +L+S LKSN + E L +G CL LGLVCLGD+ D Y+
Sbjct 590 LYALGLIHANYNTNDKKVKNYLMSQLKSN-MNDEVLQHGCCLGLGLVCLGDSNDENTYDE 648
Query 60 LRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALAL 119
L+++L+ D+AVAGE AA +GLL LGSG +ELL+Y+ +TQHEKI RACS++L
Sbjct 649 LKAILYSDSAVAGESAAYAIGLLKLGSGDDKCI--DELLAYAHDTQHEKITRACSISLGF 706
Query 120 ISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTG--KESAVKRLLHIAVSDVSD 177
+ FQ+E AD LIE+ +++ D++IRYGG FAI +AYC + +K+LLH +VSDVSD
Sbjct 707 VMFQKEREADSLIEELVSDKDAIIRYGGMFAIAMAYCGLSNCNKHIIKKLLHFSVSDVSD 766
Query 178 DVRRAA 183
DVRRAA
Sbjct 767 DVRRAA 772
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 38/181 (20%), Positives = 79/181 (43%), Gaps = 5/181 (2%)
Query 3 YGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLRS 62
Y +GL+ G+ + + L++ ++ + E + ++LG V ++ EA +
Sbjct 666 YAIGLLKLGSGDDKCIDELLA--YAHDTQHEKITRACSISLGFVMF--QKEREADSLIEE 721
Query 63 LLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALISF 122
L+ A+ G + + G +LL +S + + RA +AL +
Sbjct 722 LVSDKDAIIRYGGMFAIAMAYCGLSNCNKHIIKKLLHFSVSDVSDDVRRAAVIALGFVLC 781
Query 123 QREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRRA 182
+ + + + +RYG A A+G+A T E A+ L+ + ++D +D VR++
Sbjct 782 NSPNQVPKFLNLLIESYNPHVRYGAALALGIACSATANEEAINMLMPL-LTDTTDFVRQS 840
Query 183 A 183
A
Sbjct 841 A 841
> tpv:TP04_0424 26S proteasome regulatory subunit; K03032 26S
proteasome regulatory subunit N2
Length=1040
Score = 166 bits (421), Expect = 3e-41, Method: Composition-based stats.
Identities = 88/182 (48%), Positives = 123/182 (67%), Gaps = 4/182 (2%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
LY LGLIH + + L++ L++ E E + +G L LGLVC+G D E YE L+
Sbjct 452 LYALGLIHSNHFDASSKELLLNSLRNEGAE-ESVHHGAALGLGLVCMGQC-DYELYEELK 509
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
++F ++AV G+ AA+ +GLL+LGSG +EL ++S ETQHEKI RAC +A+A++
Sbjct 510 GVMFRNSAVPGQAAAIAIGLLMLGSGNENVV--DELYTFSYETQHEKIIRACVIAIAMVL 567
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
+QRE+ AD +I + + D++IRYGG F +AYC TG AVK+LL+ AVSDVSDDVRR
Sbjct 568 YQREKHADAIISKLCRDNDAIIRYGGMFCYAMAYCGTGSSYAVKQLLYAAVSDVSDDVRR 627
Query 182 AA 183
AA
Sbjct 628 AA 629
Score = 32.3 bits (72), Expect = 0.82, Method: Composition-based stats.
Identities = 22/97 (22%), Positives = 45/97 (46%), Gaps = 1/97 (1%)
Query 87 GAAAAAAANELLSYSKETQHEKIGRACSLALALISFQREEAADELIEQCLAEADSLIRYG 146
G ++ A +LL + + + RA ++L + ++++ A + +RYG
Sbjct 603 GTGSSYAVKQLLYAAVSDVSDDVRRAAVISLGFVLCNTPNQVPKVLKLLSASYNPHVRYG 662
Query 147 GAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRRAA 183
A+G++ +G A K +L +D S+ VR+ A
Sbjct 663 VTIALGVSCAASGPSEATK-ILQTLSTDRSEFVRQGA 698
> ath:AT2G32730 26S proteasome regulatory subunit, putative; K03032
26S proteasome regulatory subunit N2
Length=1004
Score = 166 bits (419), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 91/182 (50%), Positives = 121/182 (66%), Gaps = 7/182 (3%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
LY LGLIH I++FL L+S +E + +G CL LGL LG A D E Y+ ++
Sbjct 454 LYALGLIHANH-GEGIKQFLRDSLRSTNVEV--IQHGACLGLGLSALGTA-DEEIYDDVK 509
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
S+L+ D+AVAGE A + +GLLL+G+ A A+E+L+Y+ ETQHEKI R +L +AL
Sbjct 510 SVLYTDSAVAGEAAGISMGLLLVGT---ATEKASEMLAYAHETQHEKIIRGLALGIALTV 566
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
+ REE AD LIEQ + D +IRYGG +A+ LAY T A+++LLH AVSDVSDDVRR
Sbjct 567 YGREEGADTLIEQMTRDQDPIIRYGGMYALALAYSGTANNKAIRQLLHFAVSDVSDDVRR 626
Query 182 AA 183
A
Sbjct 627 TA 628
> ath:AT1G04810 26S proteasome regulatory subunit, putative; K03032
26S proteasome regulatory subunit N2
Length=1001
Score = 165 bits (417), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 90/182 (49%), Positives = 121/182 (66%), Gaps = 7/182 (3%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
LY LGLIH I++FL L+S +E + +G CL LGL LG A D + Y+ ++
Sbjct 454 LYALGLIHANH-GEGIKQFLRDSLRSTSVEV--IQHGACLGLGLAALGTA-DEDIYDDIK 509
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
S+L+ D+AVAGE A + +GLLL+G+ A A+E+L+Y+ ETQHEKI R +L +AL
Sbjct 510 SVLYTDSAVAGEAAGISMGLLLVGT---ATDKASEMLAYAHETQHEKIIRGLALGIALTV 566
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
+ REE AD LIEQ + D +IRYGG +A+ LAY T A+++LLH AVSDVSDDVRR
Sbjct 567 YGREEGADTLIEQMTRDQDPIIRYGGMYALALAYSGTANNKAIRQLLHFAVSDVSDDVRR 626
Query 182 AA 183
A
Sbjct 627 TA 628
> tgo:TGME49_063060 26S proteasome non-ATPase regulatory subunit
1, putative ; K03032 26S proteasome regulatory subunit N2
Length=1156
Score = 164 bits (415), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 103/182 (56%), Positives = 136/182 (74%), Gaps = 2/182 (1%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
Y LGLI P+ +R +L+ L+ EP G CL LGLVC+G+AED+E YE L+
Sbjct 544 FYALGLISANQPSTSVRDYLLEQLQVAGTASEPKQQGCCLGLGLVCMGNAEDSEVYEALK 603
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
+LFLD+AVAGE AALG+GLLLLGS + AAA +ELL+Y+++TQHEKI RAC +A+AL+
Sbjct 604 QVLFLDSAVAGEAAALGVGLLLLGS--SNAAAVDELLAYAQDTQHEKIRRACGIAVALLM 661
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
F++E+ AD LI+Q ++D+LIRYGG F I +AYC T A++RLLH++VSDVSDDVRR
Sbjct 662 FKKEQEADALIDQLCKDSDALIRYGGMFTIAMAYCATANSGAIRRLLHVSVSDVSDDVRR 721
Query 182 AA 183
AA
Sbjct 722 AA 723
> sce:YIL075C RPN2, SEN3; Rpn2p; K03032 26S proteasome regulatory
subunit N2
Length=945
Score = 160 bits (406), Expect = 2e-39, Method: Composition-based stats.
Identities = 91/191 (47%), Positives = 124/191 (64%), Gaps = 17/191 (8%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEK---------EPLINGGCLALGLVCLGDAE 52
LYGLGLI+ G R LK+ +E + L++G L +GL +G A
Sbjct 405 LYGLGLIYAG-----FGRDTTDYLKNIIVENSGTSGDEDVDVLLHGASLGIGLAAMGSA- 458
Query 53 DTEAYECLRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRA 112
+ E YE L+ +L+ D+A +GE AALG+GL +LG+G A +++ +YS+ETQH I R
Sbjct 459 NIEVYEALKEVLYNDSATSGEAAALGMGLCMLGTGKPEAI--HDMFTYSQETQHGNITRG 516
Query 113 CSLALALISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAV 172
++ LALI++ R+E AD+LI + LA +SL+RYGGAF I LAY TG SAVKRLLH+AV
Sbjct 517 LAVGLALINYGRQELADDLITKMLASDESLLRYGGAFTIALAYAGTGNNSAVKRLLHVAV 576
Query 173 SDVSDDVRRAA 183
SD +DDVRRAA
Sbjct 577 SDSNDDVRRAA 587
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 36/143 (25%), Positives = 65/143 (45%), Gaps = 3/143 (2%)
Query 41 LALGLVCLGDAEDTEAYECLRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSY 100
LA+GL + A + + +L D ++ G A + L G+G +A LL
Sbjct 517 LAVGLALINYGRQELADDLITKMLASDESLLRYGGAFTIALAYAGTGNNSAV--KRLLHV 574
Query 101 SKETQHEKIGRACSLALALISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGK 160
+ ++ + RA +AL + + +++ ++ +R G AFA+G+A G
Sbjct 575 AVSDSNDDVRRAAVIALGFVLLRDYTTVPRIVQLLSKSHNAHVRCGTAFALGIACAGKGL 634
Query 161 ESAVKRLLHIAVSDVSDDVRRAA 183
+SA+ +L D D VR+AA
Sbjct 635 QSAID-VLDPLTKDPVDFVRQAA 656
> cpv:cgd4_3950 RPN2/26s proteasome regulatory subunit ; K03032
26S proteasome regulatory subunit N2
Length=1075
Score = 155 bits (391), Expect = 9e-38, Method: Composition-based stats.
Identities = 85/183 (46%), Positives = 120/183 (65%), Gaps = 5/183 (2%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
+ LGLI+ G + + R +L+ LK NP E L +G L LGL+ G D E Y+ LR
Sbjct 476 FFALGLINAGYMDQKSREYLLEKLK-NPQRNEVLQHGASLGLGLMGFGSC-DAELYDELR 533
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
++L++D AVAGE AA +GL++ GSG +LLSY+K+TQHEKI RACS+AL +
Sbjct 534 NVLYMDNAVAGEAAAYSIGLVMFGSGCKKTIG--DLLSYAKDTQHEKIVRACSVALGFVM 591
Query 122 FQRE-EAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVR 180
+ R E ADEL +Q ++++ IRYG +GLAYC TG + A+++LL I VS++SDD +
Sbjct 592 YSRGGEEADELFQQLNSDSEHFIRYGAMHVLGLAYCGTGNQFAMEKLLQIIVSELSDDNK 651
Query 181 RAA 183
RAA
Sbjct 652 RAA 654
Score = 32.3 bits (72), Expect = 0.94, Method: Composition-based stats.
Identities = 26/102 (25%), Positives = 47/102 (46%), Gaps = 9/102 (8%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
++ LGL+ C + F + C NP + L LG+ C G E + L+
Sbjct 655 VFALGLVMCRNTQQIPQVFSLLCDSYNPHVRY----AAALTLGITCCG-MESPKVTAILK 709
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKE 103
S+ + + + A +GLGLLL+ ++N+ LS +++
Sbjct 710 SMTTDSSDIVRQAAYIGLGLLLMQQN----ESSNDKLSCTRQ 747
> xla:446265 psmd1; proteasome (prosome, macropain) 26S subunit,
non-ATPase, 1; K03032 26S proteasome regulatory subunit N2
Length=951
Score = 147 bits (370), Expect = 2e-35, Method: Composition-based stats.
Identities = 83/184 (45%), Positives = 117/184 (63%), Gaps = 10/184 (5%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLK--SNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC 59
LY LGLIH I +L++ LK SN I + +GGCL LGL LG A + Y+
Sbjct 443 LYALGLIHANHGGDIID-YLLNQLKNASNDIVR----HGGCLGLGLAALGTARQ-DVYDL 496
Query 60 LRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALAL 119
L++ L+ D AV GE A L LGL++LGS A A +++ Y++ETQHEKI R ++ +A+
Sbjct 497 LKTNLYQDDAVTGEAAGLALGLVMLGS--KNAQAIEDMVGYAQETQHEKILRGLAVGIAM 554
Query 120 ISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDV 179
+ + R E AD LIE + D ++R G + + +AYC +G A++RLLH+AVSDV+DDV
Sbjct 555 VMYGRMEEADALIESLCRDKDPILRRSGMYTVAMAYCGSGNNKAIRRLLHVAVSDVNDDV 614
Query 180 RRAA 183
RRAA
Sbjct 615 RRAA 618
> xla:734707 hypothetical protein MGC114631; K03032 26S proteasome
regulatory subunit N2
Length=951
Score = 147 bits (370), Expect = 3e-35, Method: Composition-based stats.
Identities = 83/184 (45%), Positives = 117/184 (63%), Gaps = 10/184 (5%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLK--SNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC 59
LY LGLIH I +L++ LK SN I + +GGCL LGL LG A + Y+
Sbjct 443 LYALGLIHANHGGDIID-YLLNQLKNASNDIVR----HGGCLGLGLASLGTARQ-DVYDL 496
Query 60 LRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALAL 119
L++ L+ D AV GE A L LGL++LGS A A +++ Y++ETQHEKI R ++ +A+
Sbjct 497 LKTNLYQDDAVTGEAAGLALGLVMLGS--KNAQAIEDMVGYAQETQHEKILRGLAVGIAM 554
Query 120 ISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDV 179
+ + R E AD LIE + D ++R G + + +AYC +G A++RLLH+AVSDV+DDV
Sbjct 555 VMYGRMEEADALIESLCRDKDPILRRSGMYTVAMAYCGSGNNKAIRRLLHVAVSDVNDDV 614
Query 180 RRAA 183
RRAA
Sbjct 615 RRAA 618
> mmu:70247 Psmd1, 2410026J11Rik, P112, S1; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 1; K03032 26S proteasome
regulatory subunit N2
Length=953
Score = 143 bits (360), Expect = 4e-34, Method: Composition-based stats.
Identities = 82/184 (44%), Positives = 116/184 (63%), Gaps = 10/184 (5%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLK--SNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC 59
LY LGLIH I +L++ LK SN I + +GG L LGL +G A + Y+
Sbjct 443 LYALGLIHANHGGDIID-YLLNQLKNASNDIVR----HGGSLGLGLAAMGTARQ-DVYDL 496
Query 60 LRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALAL 119
L++ L+ D AV GE A L LGL++LGS A A +++ Y++ETQHEKI R ++ +AL
Sbjct 497 LKTNLYQDDAVTGEAAGLALGLVMLGS--KNAQAIEDMVGYAQETQHEKILRGLAVGIAL 554
Query 120 ISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDV 179
+ + R E AD LIE + D ++R G + + +AYC +G A++RLLH+AVSDV+DDV
Sbjct 555 VMYGRMEEADALIESLCRDKDPILRRSGMYTVAMAYCGSGNNKAIRRLLHVAVSDVNDDV 614
Query 180 RRAA 183
RRAA
Sbjct 615 RRAA 618
> hsa:5707 PSMD1, MGC133040, MGC133041, P112, Rpn2, S1; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 1; K03032
26S proteasome regulatory subunit N2
Length=922
Score = 143 bits (360), Expect = 4e-34, Method: Composition-based stats.
Identities = 82/184 (44%), Positives = 116/184 (63%), Gaps = 10/184 (5%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLK--SNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC 59
LY LGLIH I +L++ LK SN I + +GG L LGL +G A + Y+
Sbjct 443 LYALGLIHANHGGDIID-YLLNQLKNASNDIVR----HGGSLGLGLAAMGTARQ-DVYDL 496
Query 60 LRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALAL 119
L++ L+ D AV GE A L LGL++LGS A A +++ Y++ETQHEKI R ++ +AL
Sbjct 497 LKTNLYQDDAVTGEAAGLALGLVMLGS--KNAQAIEDMVGYAQETQHEKILRGLAVGIAL 554
Query 120 ISFQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDV 179
+ + R E AD LIE + D ++R G + + +AYC +G A++RLLH+AVSDV+DDV
Sbjct 555 VMYGRMEEADALIESLCRDKDPILRRSGMYTVAMAYCGSGNNKAIRRLLHVAVSDVNDDV 614
Query 180 RRAA 183
RRAA
Sbjct 615 RRAA 618
> bbo:BBOV_II001620 18.m09940; rpn2_yeast 26S proteasome regulatory
subunit rpn2; K03032 26S proteasome regulatory subunit
N2
Length=1022
Score = 141 bits (356), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/182 (44%), Positives = 113/182 (62%), Gaps = 4/182 (2%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
LY +GLI + L +K+ +E E + +G L LGLVC+G D YE L
Sbjct 434 LYAIGLIMANHYDSEAVDILTQHIKNESVE-EAVHHGAALGLGLVCMGQC-DRAVYEELN 491
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
++L + AV+G+ AA+G+GLL+ GSG A L S +TQHEKI RAC+LA+A++
Sbjct 492 NILLKNNAVSGQAAAIGIGLLMFGSGNLEVLDA--LHSCCVDTQHEKISRACALAIAMVL 549
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
++ E+ AD +I + + D ++RYGG FA +AYC TG VK LL+ +VSDVSDDVRR
Sbjct 550 YRLEKDADSMIAKMTKDNDPVVRYGGMFAYAMAYCGTGSSKVVKALLYASVSDVSDDVRR 609
Query 182 AA 183
AA
Sbjct 610 AA 611
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 45/179 (25%), Positives = 79/179 (44%), Gaps = 9/179 (5%)
Query 4 GLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGC-LALGLVCLGDAEDTEAYECLRS 62
G+GL+ G+ N + L SC EK I+ C LA+ +V +D ++ +
Sbjct 508 GIGLLMFGSGNLEVLDALHSCCVDTQHEK---ISRACALAIAMVLYRLEKDADS--MIAK 562
Query 63 LLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALISF 122
+ + V G + G+G++ A LL S + + RA ++L +
Sbjct 563 MTKDNDPVVRYGGMFAYAMAYCGTGSSKVVKA--LLYASVSDVSDDVRRAAVISLGFVLC 620
Query 123 QREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
E ++++ +A + +RY A A+G+ C + V RLLH SD +D VR+
Sbjct 621 NTPEEVPKVLKLLVASYNPHVRYAAAIALGIT-CAASPQPDVLRLLHALSSDSADFVRQ 678
> dre:554967 psmd1, MGC55818, zgc:55818; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 1; K03032 26S proteasome
regulatory subunit N2
Length=887
Score = 128 bits (322), Expect = 8e-30, Method: Composition-based stats.
Identities = 73/182 (40%), Positives = 108/182 (59%), Gaps = 6/182 (3%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
LY LGLIH I +L+S LK+ ++ G + + Y+ L+
Sbjct 444 LYALGLIHANHGGDIID-YLLSQLKN---ASNDIVRHGGALGLGLAALGTARQDVYDLLK 499
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
S L+ D AV GE A L LGL++LGS +A A +++ Y++ETQHEKI R ++ +A++
Sbjct 500 SNLYQDDAVTGEAAGLALGLVMLGS--KSAQAIEDMVGYAQETQHEKILRGLAVGIAMVM 557
Query 122 FQREEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRR 181
+ R E AD LIE + D ++R G + + +AYC +G A++RLLH+AVSDV+DDVRR
Sbjct 558 YGRMEEADALIESLCRDKDPILRRSGMYTVAMAYCGSGNNKAIRRLLHVAVSDVNDDVRR 617
Query 182 AA 183
AA
Sbjct 618 AA 619
Score = 38.5 bits (88), Expect = 0.013, Method: Composition-based stats.
Identities = 38/180 (21%), Positives = 75/180 (41%), Gaps = 7/180 (3%)
Query 4 GLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLRSL 63
LGL+ G+ + + ++ + + E ++ G + + +V G E+ +A + SL
Sbjct 516 ALGLVMLGSKSAQAIEDMVG--YAQETQHEKILRGLAVGIAMVMYGRMEEADAL--IESL 571
Query 64 LFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALISFQ 123
+ + + GSG A LL + ++ + RA ++ I F+
Sbjct 572 CRDKDPILRRSGMYTVAMAYCGSGNNKAI--RRLLHVAVSDVNDDVRRAAVESIGFIMFR 629
Query 124 REEAADELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRRAA 183
E ++ + +RYG A A+G+ TG + A+ LL +D + VR+ A
Sbjct 630 TPEQCPSVVSLLSESYNPHVRYGAAMALGICCAGTGNKEAI-HLLEPMTNDPVNYVRQGA 688
> cel:C23G10.4 rpn-2; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-2); K03032 26S proteasome regulatory
subunit N2
Length=965
Score = 89.7 bits (221), Expect = 5e-18, Method: Composition-based stats.
Identities = 59/183 (32%), Positives = 94/183 (51%), Gaps = 6/183 (3%)
Query 2 LYGLGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLR 61
+ GLIH + L LK+ E EP+ +G CL G+ LG + YE +R
Sbjct 420 MLAYGLIHAKHGDATAMSTLAQWLKT--AENEPVRHGACLGFGVAGLGSSS-VSNYEKVR 476
Query 62 SLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
+L D AV+GE A + +GL++ +G NEL Y+ +TQH+K R LA +
Sbjct 477 EVLQRDEAVSGESAGIAMGLIM--AGHLNQEVFNELKQYTVDTQHDKTQRGIRTGLACAA 534
Query 122 FQREEAADELIEQCL-AEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVR 180
F + A+ I++ + A+++ ++R G + +AY TG V+RLL +D + DV+
Sbjct 535 FGLQGDAEPYIKEAIGAKSNPMLRSTGICMLSMAYAGTGSPDVVRRLLEKVATDPNLDVK 594
Query 181 RAA 183
R A
Sbjct 595 RYA 597
Score = 31.2 bits (69), Expect = 2.2, Method: Composition-based stats.
Identities = 39/161 (24%), Positives = 66/161 (40%), Gaps = 22/161 (13%)
Query 5 LGLIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLRSLL 64
L + + G +P + R L+ + ++P N + +G + CL +
Sbjct 565 LSMAYAGTGSPDVVRRLLEKVATDP-------NLDVKRYATIGIGFVLSKDPSTCLSYVA 617
Query 65 FLDAAVAGE---GAALGLGLLLLGSGAAAAAAANELLSYSKETQHEKIGRACSLALALIS 121
L G GAA+ LG+ G+G A A E + KE + + L+LALI
Sbjct 618 MLTEHFNGHVRYGAAMALGIACAGTGNMEAIALIEPMISDKEG---FVRKGALLSLALIM 674
Query 122 FQREE----AADELIEQCLAE-----ADSLIRYGGAFAIGL 153
Q+ + + +Q L + DSL+++G A GL
Sbjct 675 CQQTDYTCPKVNGFRKQLLKKIGEKNEDSLVKFGAIIAQGL 715
Score = 30.4 bits (67), Expect = 3.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 143 IRYGGAFAIGLAYCNTGKESAVKRLLHIAVSDVSDDVRRAA 183
+RYG A A+G+A TG A+ L+ +SD VR+ A
Sbjct 627 VRYGAAMALGIACAGTGNMEAIA-LIEPMISDKEGFVRKGA 666
> cel:T22D1.9 rpn-1; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-1); K03028 26S proteasome regulatory
subunit N1
Length=981
Score = 38.5 bits (88), Expect = 0.013, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 45/98 (45%), Gaps = 2/98 (2%)
Query 75 AALGLGLLLLGSGAAAAAAA--NELLSYSKETQHEKIGRACSLALALISFQREEAADELI 132
A L LGL+L+G+ A AA L+ S+ + R +L +ALI Q ++ +D +
Sbjct 516 AGLSLGLILVGTADAEAAMEMLQALMDKSETELTDPNMRFLALGIALIFLQTQDKSDVFV 575
Query 133 EQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLLHI 170
E + D AY TG +++LLH+
Sbjct 576 ESLRSLPDPFGAMVSTLVEVCAYAGTGNVLKIQKLLHL 613
Score = 29.6 bits (65), Expect = 6.7, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 115 LALALISFQREEAAD---ELIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVK 165
LA+ +IS ++A D L+ + S++R G +GLAY N+ +E+ K
Sbjct 434 LAIGIISSGIQDACDPASALLLDHVQSDRSIMRVGSILGLGLAYANSKRETVTK 487
> bbo:BBOV_IV009920 23.m05991; proteasome 26S regulatory subunit;
K03028 26S proteasome regulatory subunit N1
Length=1008
Score = 32.7 bits (73), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 45/98 (45%), Gaps = 6/98 (6%)
Query 75 AALGLGLLLLGSGAAAAAAA--NELLSYSKETQHEKIGRACSLA--LALISFQREEAADE 130
A+L +GL+ +G+G A+ A +LL ++ T + C +A LAL+ R EA D
Sbjct 574 ASLAIGLIFVGTGKQEASEALIQKLLDFTDNTM--DMAAKCQIACGLALLHLGRMEATDV 631
Query 131 LIEQCLAEADSLIRYGGAFAIGLAYCNTGKESAVKRLL 168
+++ A L R AY +G +R L
Sbjct 632 IVDALAAVEGDLGRIAEMMVEACAYAGSGDVLRTQRFL 669
> tgo:TGME49_066450 lysine decarboxylase domain-containing protein
Length=361
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/27 (59%), Positives = 19/27 (70%), Gaps = 1/27 (3%)
Query 48 LGDAEDTEAYECLRSLLFLDAAVAGEG 74
+ D ED EA+E LRS L D+AV GEG
Sbjct 324 ITDDED-EAFEYLRSFLLKDSAVTGEG 349
> dre:407708 lmf2b, im:7150505, im:7152257, lmf2, tmem112b, tmem153;
lipase maturation factor 2b
Length=713
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 34/126 (26%), Positives = 57/126 (45%), Gaps = 18/126 (14%)
Query 2 LYG-LGLIHCGAPNPRIRRFLISCLKSNP----------IEKEPLINGGCL-----ALGL 45
LYG G++ P+++R L+ L+++P +E + ++ CL +LG
Sbjct 33 LYGDEGILPVRLQMPKMQRPLLEQLQASPSLLWLGPSLGLEPQQVLELICLLGVLLSLGA 92
Query 46 VCLGDAEDTEAYECLRSLLFLDAAVAGEGAALGLGLLLLGSG--AAAAAAANELLSYSKE 103
V LG D+ Y CL +L V G+ +LLL +G A A L S+S+
Sbjct 93 VLLGTLRDSLTYLCLWALYLSLYNVGGDFLHSEWDVLLLEAGFLAVLVAPLGLLRSHSRH 152
Query 104 TQHEKI 109
H+ +
Sbjct 153 AFHDSL 158
> eco:b1427 rimL, ECK1420, JW1423; ribosomal-protein-L7/L12-serine
acetyltransferase (EC:2.3.1.-); K03817 ribosomal-protein-serine
acetyltransferase [EC:2.3.1.-]
Length=179
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 23/46 (50%), Gaps = 2/46 (4%)
Query 7 LIHCGAPNPRIRRFLISCLKSNPIEKEPLINGGCLALGLVCLGDAE 52
LIH A + +RRF+I C NP + + G + G CL AE
Sbjct 117 LIHHYAQSGELRRFVIKCRVDNPQSNQVALRNGFILEG--CLKQAE 160
> cel:F36D4.6 hypothetical protein
Length=134
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 3/76 (3%)
Query 41 LALGLVCLGDAE-DTEAYECLRSLLFLDAAVAGEGAALGLGLLLLGSGAAAAAAANEL-- 97
+ + L+ L AE D EA + R+ L +A + G+G L L+ S A A +NEL
Sbjct 56 IGMQLLLLRMAEKDGEAEKLARAELITEANLGGQGETTQLNPLVPVSKATAEKLSNELDE 115
Query 98 LSYSKETQHEKIGRAC 113
+ ++ + K GR C
Sbjct 116 VGRNRTARWSKSGRTC 131
> tgo:TGME49_114780 myosin G
Length=2059
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 53/122 (43%), Gaps = 7/122 (5%)
Query 22 ISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYEC---LRSLL--FLDAAVAGEGAA 76
I C+K NP K +I+ + LVC G E + LR L F+D G+
Sbjct 706 IRCIKPNPAHKPRVIHSSQILNQLVCSGVMEAIRIRKSGFALRLLHQDFVDRYRLVLGSK 765
Query 77 LGLGLLLLGSGAAAAAAANELLSYSKETQHE-KIGRACSLALALISFQREEAADE-LIEQ 134
GL L + +AA +L++ +Q E IGR A + + E A +E L+E
Sbjct 766 AAAGLRTLDAASAAQQLVTQLVANKWVSQEECLIGRTKVFAKSTVQDFLERAREEALVEP 825
Query 135 CL 136
+
Sbjct 826 VI 827
> tgo:TGME49_097820 armadillo/beta-catenin-like repeat protein
Length=512
Score = 29.3 bits (64), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 34/143 (23%), Positives = 56/143 (39%), Gaps = 28/143 (19%)
Query 22 ISCLKSNPIEKEPLINGGCLALGLVCLGDAEDTEAYECLRSLLFLDAAVAGEGAALGLGL 81
++ L S P E L + G ++L LR LL + + AAL LG
Sbjct 23 VADLSSRPQNIETLHSAGVMSL----------------LRPLLLDNVPSIQQTAALALGR 66
Query 82 LLLGSGAAAAAAANE----LLSYSKETQHEKIGRACSLALALISFQREEAADELIEQ--- 134
L S A A + L++S Q+ R+ + + +S E A +++
Sbjct 67 LASHSEELAEAVVTQEILPQLAHSVAQQNRFYKRSAAFVIRAVSKHSAELAQAVVDSGAL 126
Query 135 -----CLAEADSLIRYGGAFAIG 152
CL E D ++ A+A+G
Sbjct 127 EALTTCLEEFDPQVKESAAWAMG 149
Lambda K H
0.320 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4976880524
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40