bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eace_1846_orf5
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
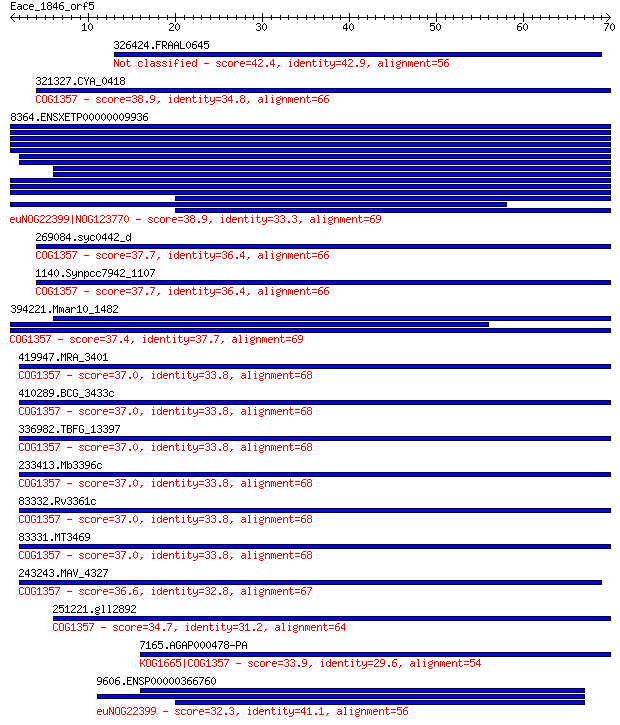
326424.FRAAL0645 42.4 0.004
321327.CYA_0418 38.9 0.043
8364.ENSXETP00000009936 38.9 0.045
269084.syc0442_d 37.7 0.097
1140.Synpcc7942_1107 37.7 0.097
394221.Mmar10_1482 37.4 0.13
419947.MRA_3401 37.0 0.16
410289.BCG_3433c 37.0 0.16
336982.TBFG_13397 37.0 0.16
233413.Mb3396c 37.0 0.16
83332.Rv3361c 37.0 0.16
83331.MT3469 37.0 0.16
243243.MAV_4327 36.6 0.25
251221.gll2892 34.7 0.79
7165.AGAP000478-PA 33.9 1.5
9606.ENSP00000366760 32.3 4.5
> 326424.FRAAL0645
Length=132
Score = 42.4 bits (98), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 25/56 (44%), Gaps = 0/56 (0%)
Query 13 CGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRLCG 68
R R RA R A R RA R A R RA RFR R A R RL CR+
Sbjct 18 VPRFRVPRFRAPRFRAPRFRAPRFRAPRFRAPRFRAPRFRAPRPRVPRLRACRVVS 73
> 321327.CYA_0418
Length=164
Score = 38.9 bits (89), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 25/76 (32%), Gaps = 10/76 (13%)
Query 4 CWLCACWLSCGRLSGGRLR----------ACRLCACRLRACRLSACRLRACRFRCCRLCA 53
+L LS L G LR RL LR L+A L L
Sbjct 57 AYLSGADLSQADLQGSNLRLANLEKAKLQGSRLQGANLRGANLTAADLSGADLSQADLAG 116
Query 54 CRLSGGRLCGCRLCGC 69
LSG L G L GC
Sbjct 117 ANLSGANLKGANLSGC 132
> 8364.ENSXETP00000009936
Length=254
Score = 38.9 bits (89), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 25/69 (36%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C AC S
Sbjct 138 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 197
Query 61 LCGCRLCGC 69
C C
Sbjct 198 YSACPYSAC 206
Score = 38.9 bits (89), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 25/69 (36%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C AC S
Sbjct 143 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 202
Query 61 LCGCRLCGC 69
C C
Sbjct 203 YSACPYSAC 211
Score = 38.9 bits (89), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 25/69 (36%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C AC S
Sbjct 148 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 207
Query 61 LCGCRLCGC 69
C C
Sbjct 208 YSACPYSAC 216
Score = 37.7 bits (86), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 25/69 (36%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C AC S
Sbjct 153 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 212
Query 61 LCGCRLCGC 69
C C
Sbjct 213 YSACPYSVC 221
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 25/69 (36%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C AC S
Sbjct 163 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSVCS 222
Query 61 LCGCRLCGC 69
C C
Sbjct 223 YSACPYSAC 231
Score = 37.4 bits (85), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 24/68 (35%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
AC AC S S AC AC AC SAC AC + C AC S
Sbjct 134 SACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPY 193
Query 62 CGCRLCGC 69
C C
Sbjct 194 SACPYSAC 201
Score = 36.6 bits (83), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
AC L A LS S AC AC AC SAC AC + C AC S
Sbjct 124 PACTLLALTLSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPY 183
Query 62 CGCRLCGC 69
C C
Sbjct 184 SACPYSAC 191
Score = 36.2 bits (82), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 24/64 (37%), Gaps = 0/64 (0%)
Query 6 LCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCR 65
L AC L LS AC AC AC SAC AC + C AC S C
Sbjct 123 LPACTLLALTLSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 182
Query 66 LCGC 69
C
Sbjct 183 YSAC 186
Score = 36.2 bits (82), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 23/64 (35%), Gaps = 0/64 (0%)
Query 6 LCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCR 65
L AC S S AC AC AC SAC AC + C AC S C
Sbjct 133 LSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACP 192
Query 66 LCGC 69
C
Sbjct 193 YSAC 196
Score = 35.8 bits (81), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 23/69 (33%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC S C AC + C C S
Sbjct 183 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSVCSYSACPYSACPYSVCSYSACP 242
Query 61 LCGCRLCGC 69
C C
Sbjct 243 YSACHYTAC 251
Score = 35.8 bits (81), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 24/69 (34%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC AC + C C S
Sbjct 168 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSVCSYSACP 227
Query 61 LCGCRLCGC 69
C C
Sbjct 228 YSACPYSVC 236
Score = 35.4 bits (80), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 24/69 (34%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ AC AC S S AC AC AC SAC C + C AC S
Sbjct 178 YSACPYSACPYSACPYSACPYSACPYSACPYSACPYSACPYSVCSYSACPYSACPYSVCS 237
Query 61 LCGCRLCGC 69
C C
Sbjct 238 YSACPYSAC 246
Score = 35.0 bits (79), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 20/50 (40%), Gaps = 0/50 (0%)
Query 20 RLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRLCGC 69
L AC L A L AC SAC AC + C AC S C C
Sbjct 122 TLPACTLLALTLSACPYSACPYSACPYSACPYSACPYSACPYSACPYSAC 171
Score = 33.9 bits (76), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 21/57 (36%), Gaps = 0/57 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLS 57
+ AC AC S S C AC AC S C AC + C AC S
Sbjct 198 YSACPYSACPYSACPYSACPYSVCSYSACPYSACPYSVCSYSACPYSACHYTACPYS 254
Score = 31.2 bits (69), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 20/50 (40%), Gaps = 0/50 (0%)
Query 20 RLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRLCGC 69
L A L AC L A LSAC AC + C AC S C C
Sbjct 117 TLLALTLPACTLLALTLSACPYSACPYSACPYSACPYSACPYSACPYSAC 166
> 269084.syc0442_d
Length=266
Score = 37.7 bits (86), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 25/66 (37%), Gaps = 0/66 (0%)
Query 4 CWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCG 63
C L L RLSG RL L L LS LR R L L+G L G
Sbjct 41 CDLQDSGLVLARLSGARLTGSNLQRANLSGADLSGADLRQTDLRGASLQRANLAGANLTG 100
Query 64 CRLCGC 69
RL G
Sbjct 101 ARLDGV 106
> 1140.Synpcc7942_1107
Length=266
Score = 37.7 bits (86), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 25/66 (37%), Gaps = 0/66 (0%)
Query 4 CWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCG 63
C L L RLSG RL L L LS LR R L L+G L G
Sbjct 41 CDLQDSGLVLARLSGARLTGSNLQRANLSGADLSGADLRQTDLRGASLQRANLAGANLTG 100
Query 64 CRLCGC 69
RL G
Sbjct 101 ARLDGV 106
> 394221.Mmar10_1482
Length=392
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 24/64 (37%), Gaps = 0/64 (0%)
Query 6 LCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCR 65
L L RLSG LR C L C LR LS L R RL L GR
Sbjct 298 LKGAELQGARLSGADLRRCNLRDCDLRGADLSGANLTHADLRGARLDPLTLGPGRAVPTN 357
Query 66 LCGC 69
L G
Sbjct 358 LSGA 361
Score = 33.1 bits (74), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 23/55 (41%), Gaps = 0/55 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACR 55
R C L C L L G RL RL R+ AC+L LR C L A +
Sbjct 56 LRDCTLTGCDLERTDLRGARLAGARLATARMLACQLDRADLRGCDLSGADLTAAQ 110
Score = 31.2 bits (69), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 26/69 (37%), Gaps = 0/69 (0%)
Query 1 FRACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGR 60
+ L L+ L LR C L C L L RL R R+ AC+L
Sbjct 36 LQGADLSGLSLAGRDLRDADLRDCTLTGCDLERTDLRGARLAGARLATARMLACQLDRAD 95
Query 61 LCGCRLCGC 69
L GC L G
Sbjct 96 LRGCDLSGA 104
> 419947.MRA_3401
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 410289.BCG_3433c
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 336982.TBFG_13397
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 233413.Mb3396c
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 83332.Rv3361c
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 83331.MT3469
Length=183
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 25/68 (36%), Gaps = 0/68 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R L + L G LR L CRLR L LR C R L R +G RL
Sbjct 80 RPLTLDDVDFTLAVLGGNDLRGLNLTGCRLRETSLVDTDLRKCVLRGADLSGARTTGARL 139
Query 62 CGCRLCGC 69
L G
Sbjct 140 DDADLRGA 147
> 243243.MAV_4327
Length=185
Score = 36.6 bits (83), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 24/67 (35%), Gaps = 0/67 (0%)
Query 2 RACWLCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRL 61
R + L G LR L CRLR L LR R L R +G RL
Sbjct 80 RPVTFDEVDFTLAVLGGNDLRGVDLSGCRLRVTSLVEADLRKAVLRGADLRGARTTGTRL 139
Query 62 CGCRLCG 68
LCG
Sbjct 140 DDADLCG 146
> 251221.gll2892
Length=457
Score = 34.7 bits (78), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 23/64 (35%), Gaps = 0/64 (0%)
Query 6 LCACWLSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCR 65
L L L G L+ L L L LR R RL C L G L GC+
Sbjct 176 LSEADLGGANLGGANLKGADLGGANLERTSLRGADLRGADLRRTRLTGCSLEGAVLEGCQ 235
Query 66 LCGC 69
+ G
Sbjct 236 VYGT 239
> 7165.AGAP000478-PA
Length=324
Score = 33.9 bits (76), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 21/54 (38%), Gaps = 0/54 (0%)
Query 16 LSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRLCGC 69
+ G LR L + +R C L+ + C L C L G L G L G
Sbjct 255 MEGVSLRVANLKSANMRNCTLNDADMAGANLESCDLTGCNLKGANLRGANLKGA 308
> 9606.ENSP00000366760
Length=248
Score = 32.3 bits (72), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 16 LSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRL 66
LS L AC L + L ACRLS+ + C L AC LS L C L
Sbjct 110 LSSMTLSACGLSSTTLFACRLSSVTVSTCSLSSVTLSACGLSRVTLSACGL 160
Score = 32.0 bits (71), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 26/56 (46%), Gaps = 0/56 (0%)
Query 11 LSCGRLSGGRLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRL 66
LS LS L AC L + L AC LS+ L ACR + C LS L C L
Sbjct 95 LSAWGLSSVTLSACCLSSMTLSACGLSSTTLFACRLSSVTVSTCSLSSVTLSACGL 150
Score = 31.2 bits (69), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 20 RLRACRLCACRLRACRLSACRLRACRFRCCRLCACRLSGGRLCGCRL 66
L AC L + + C LS+ L AC L ACRLS L L
Sbjct 174 TLSACHLSSMTVSTCGLSSMTLFACGLSSMTLSACRLSNVTLSNFPL 220
Lambda K H
0.346 0.149 0.654
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22781900540
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40