bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eace_1954_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
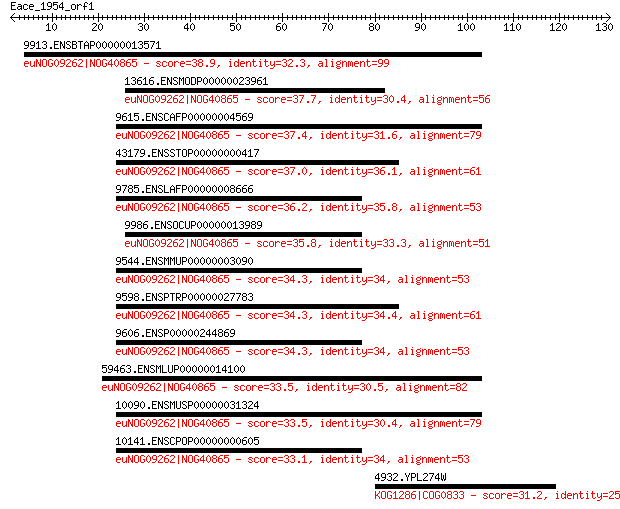
9913.ENSBTAP00000013571 38.9 0.043
13616.ENSMODP00000023961 37.7 0.10
9615.ENSCAFP00000004569 37.4 0.12
43179.ENSSTOP00000000417 37.0 0.16
9785.ENSLAFP00000008666 36.2 0.26
9986.ENSOCUP00000013989 35.8 0.41
9544.ENSMMUP00000003090 34.3 1.0
9598.ENSPTRP00000027783 34.3 1.1
9606.ENSP00000244869 34.3 1.1
59463.ENSMLUP00000014100 33.5 1.9
10090.ENSMUSP00000031324 33.5 2.0
10141.ENSCPOP00000000605 33.1 2.7
4932.YPL274W 31.2 8.7
> 9913.ENSBTAP00000013571
Length=170
Score = 38.9 bits (89), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 44/110 (40%), Gaps = 11/110 (10%)
Query 4 ARLAPLMWLISAPVFDFL--CTSRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAY 61
AR P + L+ VF L S T P C+P D+ A V EDNPR VS+
Sbjct 11 ARQVPTLLLVLEIVFYLLQAVFSTTVIPSCIPGESGDNCTALVQTEDNPRVAQVSITKCS 70
Query 62 EEFVPNTLSGRLVLL---------CQVRRSGLHAMDENLDRKPSLSSSVV 102
+ L G+ + L C+V +G+ L + LS V
Sbjct 71 ADMNGYCLHGQCIYLVDMSENYCRCEVGYTGVRCEHFFLTVQKPLSKEYV 120
> 13616.ENSMODP00000023961
Length=162
Score = 37.7 bits (86), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 26 TNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLLCQVRR 81
T P C+P DD+ A V +E+NPR V++++ E L G+ + L +++
Sbjct 27 TVIPSCIPGESDDNCTALVQIENNPRVAQVAIMNCGENMQGYCLHGQCIYLVDMKQ 82
> 9615.ENSCAFP00000004569
Length=148
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 38/88 (43%), Gaps = 9/88 (10%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL------- 76
S T P C+P +D+ A V +EDNPR VS++ + L G+ + L
Sbjct 11 STTVIPSCMPGESEDNCTALVQIEDNPRVAQVSIIKCGSDMNGYCLHGQCIYLVDMSQTY 70
Query 77 --CQVRRSGLHAMDENLDRKPSLSSSVV 102
C+V +G+ L + LS V
Sbjct 71 CRCEVGYTGVRCEHFYLTVQQPLSKEYV 98
> 43179.ENSSTOP00000000417
Length=119
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 9/70 (12%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL------- 76
S T P C+P DD+ A V +EDNPR VS+ + L G+ + L
Sbjct 9 STTVIPSCIPGESDDNCTALVQMEDNPRVAQVSITKCGTDMNGYCLHGQCIYLVDMSENY 68
Query 77 --CQVRRSGL 84
C+V +G+
Sbjct 69 CRCEVGYTGV 78
> 9785.ENSLAFP00000008666
Length=169
Score = 36.2 bits (82), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL 76
S T P C+P DD+ A V EDNPR VS+ + L G + L
Sbjct 32 STTVIPSCIPGESDDNCTALVQTEDNPRVAQVSITKCGSDMNGYCLHGHCIFL 84
> 9986.ENSOCUP00000013989
Length=169
Score = 35.8 bits (81), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 26 TNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL 76
T P C+P +D+ A V +EDNPR VS+ + L G+ + L
Sbjct 34 TVIPSCIPGEAEDNCTALVQMEDNPRVAQVSITKCGSDMNGYCLHGQCIYL 84
> 9544.ENSMMUP00000003090
Length=169
Score = 34.3 bits (77), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL 76
S T P C+P D+ A V EDNPR VS+ + L G+ + L
Sbjct 32 STTVIPSCIPGESSDNCTALVQTEDNPRVAQVSITKCNSDMNGYCLHGQCIYL 84
> 9598.ENSPTRP00000027783
Length=169
Score = 34.3 bits (77), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 30/70 (42%), Gaps = 9/70 (12%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL------- 76
S T P C+P D+ A V EDNPR VS+ + L G+ + L
Sbjct 32 STTVIPSCIPGESSDNCTALVQTEDNPRVAQVSITKCSSDMNGYCLHGQCIYLVDMSQNY 91
Query 77 --CQVRRSGL 84
C+V +G+
Sbjct 92 CRCEVGYTGV 101
> 9606.ENSP00000244869
Length=169
Score = 34.3 bits (77), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL 76
S T P C+P D+ A V EDNPR VS+ + L G+ + L
Sbjct 32 STTVIPSCIPGESSDNCTALVQTEDNPRVAQVSITKCSSDMNGYCLHGQCIYL 84
> 59463.ENSMLUP00000014100
Length=146
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 36/91 (39%), Gaps = 9/91 (9%)
Query 21 LCTSRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL---- 76
L T P C+P +D+ A V E+NPR VS+ + L G+ + L
Sbjct 6 LVLGTTVIPSCIPGESEDNCTALVKTENNPRVAQVSITKCSSDMNGYCLHGQCIFLVDMS 65
Query 77 -----CQVRRSGLHAMDENLDRKPSLSSSVV 102
C+V +GL L + LS V
Sbjct 66 EKYCRCEVGYAGLRCEHFFLTVQQPLSKEYV 96
> 10090.ENSMUSP00000031324
Length=162
Score = 33.5 bits (75), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 35/88 (39%), Gaps = 9/88 (10%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL------- 76
S T P C+P +D+ A V +ED+PR V + + L G+ + L
Sbjct 25 STTVIPSCIPGESEDNCTALVQMEDDPRVAQVQITKCSSDMDGYCLHGQCIYLVDMREKF 84
Query 77 --CQVRRSGLHAMDENLDRKPSLSSSVV 102
C+V +GL L LS V
Sbjct 85 CRCEVGYTGLRCEHFFLTVHQPLSKEYV 112
> 10141.ENSCPOP00000000605
Length=162
Score = 33.1 bits (74), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 1/53 (1%)
Query 24 SRTNFPECVPWAGDDDWCADVGLEDNPRFLPVSLLHAYEEFVPNTLSGRLVLL 76
S T P C+P D + A V +EDNPR VS+ + L G+ +L+
Sbjct 28 STTVIPSCIPGESDSNCTALVQMEDNPRA-QVSITKCASDMQGYCLHGQCILV 79
> 4932.YPL274W
Length=587
Score = 31.2 bits (69), Expect = 8.7, Method: Composition-based stats.
Identities = 10/39 (25%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 80 RRSGLHAMDENLDRKPSLSSSVVKWPTKKQPYENMLARK 118
++S LH + E+L+ ++W QPY+ +L+++
Sbjct 42 KQSNLHVIPEDLENSEQTEQEKIQWKLASQPYQKVLSQR 80
Lambda K H
0.320 0.133 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 23020010250
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40