bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eace_2683_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
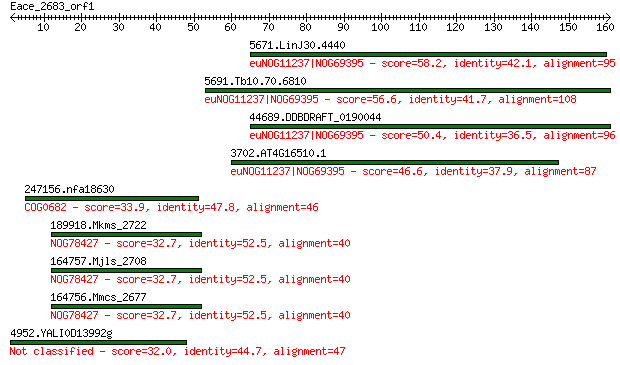
5671.LinJ30.4440 58.2 9e-08
5691.Tb10.70.6810 56.6 2e-07
44689.DDBDRAFT_0190044 50.4 2e-05
3702.AT4G16510.1 46.6 2e-04
247156.nfa18630 33.9 1.8
189918.Mkms_2722 32.7 3.6
164757.Mjls_2708 32.7 3.6
164756.Mmcs_2677 32.7 3.6
4952.YALI0D13992g 32.0 6.4
> 5671.LinJ30.4440
Length=270
Score = 58.2 bits (139), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 40/99 (40%), Positives = 54/99 (54%), Gaps = 9/99 (9%)
Query 65 SSYYSVCDLKERLPMLTANSVCQLCKTIVVENTKHAGPE--DRLNSRYYLVVLQSQDKLN 122
S+YY +L+ R L A ++ LCKT+++ENT + DR NSRYYLVV Q D+ N
Sbjct 103 SNYYQE-NLQWRRDALRAPTIRHLCKTVLMENTHCTNKDCADRNNSRYYLVVYQYVDRFN 161
Query 123 GERVKDLIKAENAKVGRRLGNKKLNFNFCQSFEA--LTG 159
+ V I+ N +G+ KK NF EA LTG
Sbjct 162 SDMVGRAIQELNPGMGK----KKFNFRLADPAEAEKLTG 196
> 5691.Tb10.70.6810
Length=261
Score = 56.6 bits (135), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 45/113 (39%), Positives = 55/113 (48%), Gaps = 11/113 (9%)
Query 53 HRCVFRLFRINCSSYYSVCDLKERLPMLTANSVCQLCKTIVVENTKHAGPED---RLNSR 109
H +FR S YY L+ R L A SV LCK+I++ENT H +D R NSR
Sbjct 82 HSLTTAVFRWVPSDYYQ-HTLQWRRDELAAPSVHHLCKSILMENT-HCTNKDCGVRENSR 139
Query 110 YYLVVLQSQDKLNGERVKDLIKAENAKVGRRLGNKKLNFNFCQSFEA--LTGF 160
YYL+V +K + E V +K N LG KK NF A LTGF
Sbjct 140 YYLIVYPYTEKFDAEMVMRHVKGLNEG----LGKKKFNFRLAAPDVALQLTGF 188
> 44689.DDBDRAFT_0190044
Length=230
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 51/98 (52%), Gaps = 7/98 (7%)
Query 65 SSYYSVCDLKERLPMLTANSVCQLCKTIVVENTKHAGPEDRL--NSRYYLVVLQSQDKLN 122
S YY+ L +R L A S L K+I++ENT+ + + NSRYY V++Q K+
Sbjct 58 SPYYT-WTLDQRASYLDAFSPEYLLKSIILENTECTNTDTSIPTNSRYYCVIIQYTTKIQ 116
Query 123 GERVKDLIKAENAKVGRRLGNKKLNFNFCQSFEALTGF 160
++ IK N+ KK +F QS E+LTGF
Sbjct 117 SHKIFKFIKNMNSNESA----KKFHFTLAQSAESLTGF 150
> 3702.AT4G16510.1
Length=232
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 51/92 (55%), Gaps = 6/92 (6%)
Query 60 FRINCSSYYSVCDLKERLPMLTANSVCQLCKTIVVENTKHAGP----EDRLNSRYYLVVL 115
F+ + YY L+ R +L A+SV LCK+IV+ NT+ A D NS+YY+VV+
Sbjct 70 FKRVATDYYD-WPLESRRDVLGASSVDHLCKSIVLVNTQAASNILDCSDPNNSKYYVVVV 128
Query 116 QSQDKLNGERVKDLIKAEN-AKVGRRLGNKKL 146
Q + N E VK + + N K+ ++ N +L
Sbjct 129 QYTARFNAEAVKQFLYSLNEGKIPKKRFNLRL 160
> 247156.nfa18630
Length=491
Score = 33.9 bits (76), Expect = 1.8, Method: Composition-based stats.
Identities = 22/50 (44%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query 5 AADKSAA----AADEATTDTSTADTAAEAAATDAPAEAAAAAAAAGAASG 50
AA+KSAA AA EA DTS AD A + A + A +A +GA G
Sbjct 403 AAEKSAADKESAAGEAAADTSAADQPAADKSGSAKSAADKSAGKSGAGRG 452
> 189918.Mkms_2722
Length=847
Score = 32.7 bits (73), Expect = 3.6, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query 12 AADEATTDTSTADTAAEAAA-TDAPAEAAAAA--AAAGAASGE 51
AA+ ATTD STA T+A A A + AP+E A + A G SG+
Sbjct 782 AAETATTDPSTASTSAPATAESSAPSEPTATSDPTADGTTSGD 824
> 164757.Mjls_2708
Length=844
Score = 32.7 bits (73), Expect = 3.6, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query 12 AADEATTDTSTADTAAEAAA-TDAPAEAAAAA--AAAGAASGE 51
AA+ ATTD STA T+A A A + AP+E A + A G SG+
Sbjct 779 AAETATTDPSTASTSAPATAESSAPSEPTATSDPTADGTTSGD 821
> 164756.Mmcs_2677
Length=847
Score = 32.7 bits (73), Expect = 3.6, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query 12 AADEATTDTSTADTAAEAAA-TDAPAEAAAAA--AAAGAASGE 51
AA+ ATTD STA T+A A A + AP+E A + A G SG+
Sbjct 782 AAETATTDPSTASTSAPATAESSAPSEPTATSDPTADGTTSGD 824
> 4952.YALI0D13992g
Length=906
Score = 32.0 bits (71), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 1 AIAPAADKSAAAADEATTDTSTADTAAEAAATDAPAEAAAAAAAAGA 47
AI P+AD + +A TT+T AD A A+T APA A A+ A A
Sbjct 430 AITPSADSTGSAVGAITTETPVAD--APTASTTAPAPATIASTATPA 474
Lambda K H
0.315 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 26871144147
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40