bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eace_3363_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
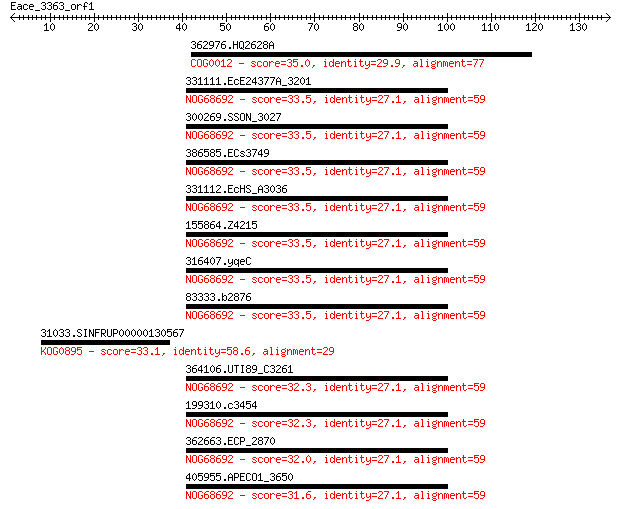
362976.HQ2628A 35.0 0.65
331111.EcE24377A_3201 33.5 1.8
300269.SSON_3027 33.5 1.8
386585.ECs3749 33.5 1.9
331112.EcHS_A3036 33.5 1.9
155864.Z4215 33.5 1.9
316407.yqeC 33.5 1.9
83333.b2876 33.5 1.9
31033.SINFRUP00000130567 33.1 2.2
364106.UTI89_C3261 32.3 4.1
199310.c3454 32.3 4.1
362663.ECP_2870 32.0 5.1
405955.APECO1_3650 31.6 6.4
> 362976.HQ2628A
Length=390
Score = 35.0 bits (79), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 35/77 (45%), Gaps = 9/77 (11%)
Query 42 PDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLAIPSPKNPLFYLNRFYNSANPF 101
PDDPQ W S QM L + +P+ L+A + P+ +NR ++++ P
Sbjct 191 PDDPQQW---------SMDDQMALAVDIRERTKPIVLVANKADIAPVENINRLHDTSKPV 241
Query 102 CAAMMQQEKAAAAAAAA 118
A E+A AA A
Sbjct 242 VPATADGERALRRAADA 258
> 331111.EcE24377A_3201
Length=235
Score = 33.5 bits (75), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 140 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 199
> 300269.SSON_3027
Length=235
Score = 33.5 bits (75), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 140 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 199
> 386585.ECs3749
Length=235
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 140 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 199
> 331112.EcHS_A3036
Length=256
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 161 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 220
> 155864.Z4215
Length=235
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 140 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 199
> 316407.yqeC
Length=256
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 161 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 220
> 83333.b2876
Length=256
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 161 STENVHRWSQFADITGLTPDATLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 220
> 31033.SINFRUP00000130567
Length=433
Score = 33.1 bits (74), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 8 FGLISIDTSTAADSLSLPAL--FVRVYWYKE 36
FGL DT +AAD+ S P + FVRV WY E
Sbjct 58 FGLCGSDTESAADTPSRPLVRGFVRVQWYPE 88
> 364106.UTI89_C3261
Length=256
Score = 32.3 bits (72), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 161 STENVHRWSQFADITGLTPDAPLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 220
> 199310.c3454
Length=256
Score = 32.3 bits (72), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 161 STENVHRWSQFADITGLTPDAPLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 220
> 362663.ECP_2870
Length=183
Score = 32.0 bits (71), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 88 STENVHRWSQFADITGLTPDAPLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 147
> 405955.APECO1_3650
Length=200
Score = 31.6 bits (70), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 41 SPDDPQDWEPHAEITGVSSQQQMPLQELLPFVERPLGLLA-IPSPKNPLFYLNRFYNSAN 99
S ++ W A+ITG++ + L +L+ V P G +P ++++NRF N
Sbjct 105 STENVHRWSQFADITGLTPDAPLQLSDLVALVRHPQGAFKNVPQGCRRVWFINRFSQCEN 164
Lambda K H
0.315 0.129 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22513421946
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40