bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Emax_0577_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
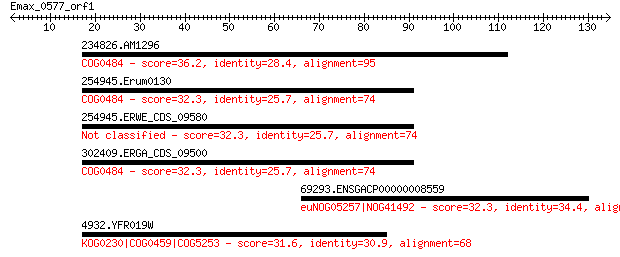
234826.AM1296 36.2 0.30
254945.Erum0130 32.3 4.2
254945.ERWE_CDS_09580 32.3 4.2
302409.ERGA_CDS_09500 32.3 4.5
69293.ENSGACP00000008559 32.3 4.5
4932.YFR019W 31.6 7.8
> 234826.AM1296
Length=379
Score = 36.2 bits (82), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 43/103 (41%), Gaps = 16/103 (15%)
Query 17 GASARSGGETGDAAAKLAPSLSLKLHRASARRGSDLHSGIPAKRLMLQVPTEKGGRANMP 76
G + G + GD L + +K H+ RR SDLH +P K + + GG MP
Sbjct 236 GEAGFRGAQEGD----LYVYIRVKEHKFFTRRSSDLHCSVPIKMTIAAL----GGEIEMP 287
Query 77 AYPSESSEPQTPVG--------FGGPTSPDVWLSDSALNLYLH 111
+ ++ + P G G P+V D ++Y+H
Sbjct 288 SIDGSWTKLKIPEGTQSGDQIRMRGKGMPEVNSKDRRGDMYVH 330
> 254945.Erum0130
Length=382
Score = 32.3 bits (72), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 35/74 (47%), Gaps = 8/74 (10%)
Query 17 GASARSGGETGDAAAKLAPSLSLKLHRASARRGSDLHSGIPAKRLMLQVPTEKGGRANMP 76
G + G ++GD L ++K H+ R G+DL+ +P K ++ + GG MP
Sbjct 239 GEAGYRGAQSGD----LYVYPNIKKHKFFTRNGADLYCNVPIKMILATL----GGHIEMP 290
Query 77 AYPSESSEPQTPVG 90
+ ++ + P G
Sbjct 291 SIDGTWTKVKVPEG 304
> 254945.ERWE_CDS_09580
Length=382
Score = 32.3 bits (72), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 35/74 (47%), Gaps = 8/74 (10%)
Query 17 GASARSGGETGDAAAKLAPSLSLKLHRASARRGSDLHSGIPAKRLMLQVPTEKGGRANMP 76
G + G ++GD L ++K H+ R G+DL+ +P K ++ + GG MP
Sbjct 239 GEAGYRGAQSGD----LYVYPNIKKHKFFTRNGADLYCNVPIKMILATL----GGHIEMP 290
Query 77 AYPSESSEPQTPVG 90
+ ++ + P G
Sbjct 291 SIDGTWTKVKVPEG 304
> 302409.ERGA_CDS_09500
Length=382
Score = 32.3 bits (72), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 35/74 (47%), Gaps = 8/74 (10%)
Query 17 GASARSGGETGDAAAKLAPSLSLKLHRASARRGSDLHSGIPAKRLMLQVPTEKGGRANMP 76
G + G ++GD L ++K H+ R G+DL+ +P K ++ + GG MP
Sbjct 239 GEAGYRGAQSGD----LYVYPNIKKHKFFTRNGADLYCNVPIKMILATL----GGHIEMP 290
Query 77 AYPSESSEPQTPVG 90
+ ++ + P G
Sbjct 291 SIDGTWTKVKVPEG 304
> 69293.ENSGACP00000008559
Length=524
Score = 32.3 bits (72), Expect = 4.5, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 34/68 (50%), Gaps = 9/68 (13%)
Query 66 PTEKGGRANMPAYPSESSEPQTPVGFGGPTSPDVWLSDSALNLYLHYNKI-DSDDSM--- 121
P E GG N+ A +S+ T V FG DVWL N++L +KI ++ D +
Sbjct 91 PVEAGGPYNVTAAAQDSTAALTDVLFG-----DVWLCGGQSNMWLQTSKIFNASDELRIA 145
Query 122 ESLPSVKV 129
E P +++
Sbjct 146 EKYPHIRI 153
> 4932.YFR019W
Length=2278
Score = 31.6 bits (70), Expect = 7.8, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 8/74 (10%)
Query 17 GASARSGG------ETGDAAAKLAPSLSLKLHRASARRGSDLHSGIPAKRLMLQVPTEKG 70
A A++GG +TGD + ++PS+S H R S++ S + K +L P +G
Sbjct 1901 AAPAKTGGNSGGTTQTGDPSVNISPSVSTTSHNKG--RDSEISSLVTTKEGLLNTPPIEG 1958
Query 71 GRANMPAYPSESSE 84
R P S+
Sbjct 1959 ARDRTPQESQTHSQ 1972
Lambda K H
0.308 0.125 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22682284714
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40