bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_0010_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
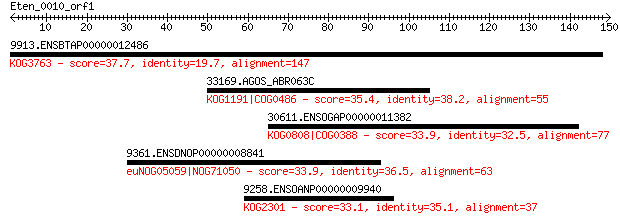
9913.ENSBTAP00000012486 37.7 0.11
33169.AGOS_ABR063C 35.4 0.48
30611.ENSOGAP00000011382 33.9 1.3
9361.ENSDNOP00000008841 33.9 1.3
9258.ENSOANP00000009940 33.1 2.4
> 9913.ENSBTAP00000012486
Length=620
Score = 37.7 bits (86), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/149 (19%), Positives = 54/149 (36%), Gaps = 6/149 (4%)
Query 1 GSGTDDGASEAHQLRVLLPQ-RPYDRLWLFRQVHQAVERDHWPVHGHFEGPRGACLPTGL 59
G+GT + + ++ +P R YD+ WL + P+ H+E R
Sbjct 108 GAGTSQDGTSKNWFKITIPYGRKYDKSWLLNVIQSKCSVPFTPIEFHYENTRAQFFVEDA 167
Query 60 GSRQPFAAVRKRGQHFEGVGRGAAATQRRPPRPRRRSLRHRLGP-QRRRCSFGGQRPHAK 118
+ AV + E PP S+++ L P Q + + +
Sbjct 168 STASALKAVNYKILDRENRRISIIINSSSPP----HSVQNELKPEQVEQLKLIMSKRYDG 223
Query 119 NLEALKSRGAAAAAAAAAAGVEVCRHRSS 147
+ +AL +G + A ++V +R S
Sbjct 224 SQQALDLKGLRSDPDLVAQNIDVVLNRRS 252
> 33169.AGOS_ABR063C
Length=493
Score = 35.4 bits (80), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query 50 PRGACLPTGLGSRQPFAAVRKRGQHFEGVGRGAAATQRRPPRPRRRSLRHRLGPQ 104
P L T +G + A VR G H V + +RPP PRR SLR+ PQ
Sbjct 19 PTIYALSTPMGQKSAIAVVRVSGTHARLVYEKLTDS-KRPPTPRRASLRNLYSPQ 72
> 30611.ENSOGAP00000011382
Length=384
Score = 33.9 bits (76), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 31/77 (40%), Gaps = 6/77 (7%)
Query 65 FAAVRKRGQHFEGVGRGAAATQRRPPRPRRRSLRHRLGPQRRRCSFGGQRPHAKNLEALK 124
F A K +G AA Q RPPR R +G + R P A+ + AL
Sbjct 44 FEAASKEDFELQGYAFEAAEEQLRPPRITR------VGLVQNRTPLPADTPVAEQVSALH 97
Query 125 SRGAAAAAAAAAAGVEV 141
R A AAA GV +
Sbjct 98 RRIEAIVEVAAACGVNI 114
> 9361.ENSDNOP00000008841
Length=743
Score = 33.9 bits (76), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 30/63 (47%), Gaps = 9/63 (14%)
Query 30 RQVHQAVERDHWPVHGHFEGPRGACLPTGLGSRQPFAAVRKRGQHFEGVGRGAAATQRRP 89
R H+ + ++ P H + P CLP QP + RG EG GRG T+RRP
Sbjct 193 RHSHEKPDSENIPPSKHHQQPPHNCLP------QPEPEAKTRG---EGDGRGCEKTERRP 243
Query 90 PRP 92
RP
Sbjct 244 ERP 246
> 9258.ENSOANP00000009940
Length=1974
Score = 33.1 bits (74), Expect = 2.4, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 59 LGSRQPFAAVRKRGQHFEGVGRGAAATQRRPPRPRRR 95
+G R F A+ + GQH + + QRR PRP +
Sbjct 1452 IGGRPDFTAIEQVGQHSSSLQPQVTSPQRRTPRPENK 1488
Lambda K H
0.322 0.135 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22854363588
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40