bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_2684_orf1
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
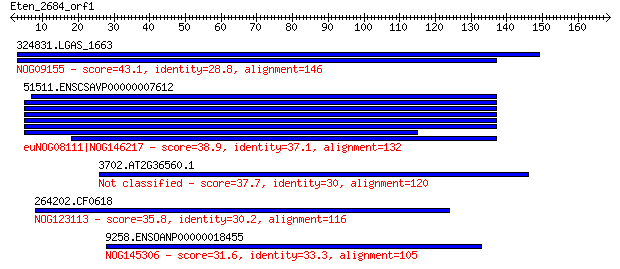
324831.LGAS_1663 43.1 0.003
51511.ENSCSAVP00000007612 38.9 0.066
3702.AT2G36560.1 37.7 0.15
264202.CF0618 35.8 0.57
9258.ENSOANP00000018455 31.6 9.0
> 324831.LGAS_1663
Length=2449
Score = 43.1 bits (100), Expect = 0.003, Method: Composition-based stats.
Identities = 42/153 (27%), Positives = 64/153 (41%), Gaps = 20/153 (13%)
Query 3 TEDEELAGVDSPSPLTEPLEEKRTGPVETPVQVG-------PVTEPAPEEEAESTEDEEL 55
T+ EE D P+ TEP + + + P Q P+T P + +E T+ EE
Sbjct 2216 TKPEEPTTPDKPARPTEPTKPEEPTTPDKPAQPSEPTKPEEPITPDKPAQPSEPTKPEEP 2275
Query 56 VGVDGPSPPTLPLEEKRTGPVETPVQVGPLTTPTPEEEAESTEGEELAGVDSPSPLTEPL 115
D P+ PT P T P E P T P + +E + EE D P+ +EP
Sbjct 2276 TTPDKPARPTEP-----TKPEE------PTTPDKPAQPSEPIKPEEPTTPDKPAQPSEPT 2324
Query 116 EEKRTGPVETPVQVGPLEAPKADAEASADTDDE 148
E K + P Q P E K + + ++ ++
Sbjct 2325 EPKEPTTPDKPTQ--PSEPTKPEEQTISNKSNQ 2355
Score = 38.5 bits (88), Expect = 0.078, Method: Composition-based stats.
Identities = 35/141 (24%), Positives = 51/141 (36%), Gaps = 7/141 (4%)
Query 3 TEDEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTEDEELVGVDGPS 62
+ D E + P+ +K P E P T P E T+ EE D P+
Sbjct 2187 SNDIERTVIYKAKPVEPTTPDKPAQPSEPTKPEEPTTPDKPARPTEPTKPEEPTTPDKPA 2246
Query 63 PPTLPLE-------EKRTGPVETPVQVGPLTTPTPEEEAESTEGEELAGVDSPSPLTEPL 115
P+ P + +K P E P T P E T+ EE D P+ +EP+
Sbjct 2247 QPSEPTKPEEPITPDKPAQPSEPTKPEEPTTPDKPARPTEPTKPEEPTTPDKPAQPSEPI 2306
Query 116 EEKRTGPVETPVQVGPLEAPK 136
+ + + P Q PK
Sbjct 2307 KPEEPTTPDKPAQPSEPTEPK 2327
> 51511.ENSCSAVP00000007612
Length=916
Score = 38.9 bits (89), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 49/136 (36%), Positives = 66/136 (48%), Gaps = 15/136 (11%)
Query 7 ELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTE---DEELVGVDGPSP 63
L G+ P P TEP K +GP + +G +T P P E A+ + + L G+ GP P
Sbjct 755 NLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGFNFNLGGLTGPKP 811
Query 64 PTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEG--EELAGVDSPSPLTEPLEEKRT 120
T P K +GP + +G LT P P E A+ S G L G+ P P TEP K +
Sbjct 812 -TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGFNFNLGGLTGPKP-TEP--AKPS 865
Query 121 GPVETPVQVGPLEAPK 136
GP + +G L PK
Sbjct 866 GPGGFNINLGGLTGPK 881
Score = 37.4 bits (85), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 65/140 (46%), Gaps = 17/140 (12%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAE----STEDEELVGVDG 60
+ L G+ P P TEP K +GP + +G +T P P E A+ + L G+ G
Sbjct 562 NINLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLTG 618
Query 61 PSPPTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPLE 116
P P T P K +GP + +G LT P P E A+ S G L G+ P P TEP
Sbjct 619 PKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLSGPKP-TEP-- 672
Query 117 EKRTGPVETPVQVGPLEAPK 136
K +GP +G L PK
Sbjct 673 AKPSGPGGFNFNLGGLTGPK 692
Score = 36.2 bits (82), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 45/137 (32%), Positives = 64/137 (46%), Gaps = 11/137 (8%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTEDEEL-VGVDGPSP 63
+ L G+ P P TEP K +GP + +G +T P P E A+ + + + GP+
Sbjct 466 NINLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGPTG 522
Query 64 PTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPLEEKR 119
P K +GP + +G LT P P E A+ S G L G+ P P TEP K
Sbjct 523 PKPTEPAKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLTGPKP-TEP--AKP 579
Query 120 TGPVETPVQVGPLEAPK 136
+GP + +G L PK
Sbjct 580 SGPGGFNINLGGLTGPK 596
Score = 36.2 bits (82), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 49/141 (34%), Positives = 66/141 (46%), Gaps = 21/141 (14%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTE---DEELVGVDGP 61
+ L G+ P P TEP K +GP + +G +T P P E A+ + + L G+ GP
Sbjct 682 NFNLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGFNFNLGGLTGP 738
Query 62 SP--PTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPL 115
P PT K +GP + +G LT P P E A+ S G L G+ P P TEP
Sbjct 739 KPTEPT-----KPSGPGGFTINLGGLTGPKPTEPAKPSGPGGFNINLGGLTGPKP-TEP- 791
Query 116 EEKRTGPVETPVQVGPLEAPK 136
K +GP +G L PK
Sbjct 792 -AKPSGPGFN-FNLGGLTGPK 810
Score = 35.8 bits (81), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 65/140 (46%), Gaps = 17/140 (12%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAE----STEDEELVGVDG 60
+ L G P P TEP K +GP + +G +T P P E A+ + L G+ G
Sbjct 514 NINLGGPTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLTG 570
Query 61 PSPPTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPLE 116
P P T P K +GP + +G LT P P E A+ S G L G+ P P TEP
Sbjct 571 PKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLTGPKP-TEP-- 624
Query 117 EKRTGPVETPVQVGPLEAPK 136
K +GP + +G L PK
Sbjct 625 AKPSGPGGFNINLGGLTGPK 644
Score = 33.9 bits (76), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 64/140 (45%), Gaps = 18/140 (12%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAE----STEDEELVGVDG 60
+ L G+ P P TEP K +GP + +G +T P P E A+ + L G+ G
Sbjct 610 NINLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGGFNINLGGLSG 666
Query 61 PSPPTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPLE 116
P P T P K +GP +G LT P P E A+ S G L G+ P P TEP
Sbjct 667 PKP-TEP--AKPSGPGGFNFNLGGLTGPKPTEPAKPSGPGGFNINLGGLTGPKP-TEP-- 720
Query 117 EKRTGPVETPVQVGPLEAPK 136
K +GP +G L PK
Sbjct 721 AKPSGPGFN-FNLGGLTGPK 739
Score = 33.5 bits (75), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 57/116 (49%), Gaps = 13/116 (11%)
Query 5 DEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTE---DEELVGVDGP 61
+ L G+ P P TEP K +GP + +G +T P P E A+ + + L G+ GP
Sbjct 800 NFNLGGLTGPKP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGFNFNLGGLTGP 856
Query 62 SPPTLPLEEKRTGPVETPVQVGPLTTPTPEEEAE-STEG--EELAGVDSPSPLTEP 114
P T P K +GP + +G LT P P E A+ S G L G+ P P TEP
Sbjct 857 KP-TEP--AKPSGPGGFNINLGGLTGPKPTEPAKPSGPGFNFNLGGLTGPKP-TEP 908
Score = 32.0 bits (71), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 43/124 (34%), Positives = 59/124 (47%), Gaps = 10/124 (8%)
Query 18 TEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTEDEEL-VGVDGPSPPTLPLEEKRTGPV 76
TEP K +GP E + +G T P P E A+ + EL + + GP+ P K +GP
Sbjct 382 TEP--AKPSGPGELNINLGGPTGPKPSEPAKPSGPGELNINLGGPTGPKPTEPAKPSGPG 439
Query 77 ETPVQVGPLTTPTPEEEAE-STEGE---ELAGVDSPSPLTEPLEEKRTGPVETPVQVGPL 132
+ +G T P P E A+ S G L G+ P P TEP K +GP + +G L
Sbjct 440 GFNINLGGPTGPKPTEPAKPSGPGGFNINLGGLTGPKP-TEP--AKPSGPGGFNINLGGL 496
Query 133 EAPK 136
PK
Sbjct 497 TGPK 500
> 3702.AT2G36560.1
Length=574
Score = 37.7 bits (86), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 54/120 (45%), Gaps = 12/120 (10%)
Query 26 TGPVETPVQVGPVTEPAPEEEAESTEDEELVGVDGPSPPTLPLEEKRTGPVETPVQVGPL 85
T PV+ P VGP + ++E + V +PP L TGPV+ P VGP
Sbjct 255 TRPVQQPEMVGP-----SHSQKRNSESNRTMVVLSATPPNT-LASSSTGPVQQPEMVGP- 307
Query 86 TTPTPEEEAESTEGEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPLEAPKADAEASADT 145
+ ++E + V S +P L TGPV+ P VGP + K ++E++ T
Sbjct 308 ----SHSQKRNSESNKTMVVLSATP-PNTLASSSTGPVQQPEMVGPSHSQKRNSESNRTT 362
> 264202.CF0618
Length=816
Score = 35.8 bits (81), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 56/116 (48%), Gaps = 20/116 (17%)
Query 8 LAGVDSPSPLTEPLEEKRTGPVETPVQVGPVTEPAPEEEAESTEDEELVGVDGPSPPTLP 67
L+G +PSP+T+P+ + VQ PV EP+ E ++ L G PSP T P
Sbjct 166 LSGGTTPSPVTQPMSQ---------VQAAPVPEPSMNPEMQALL-RLLSGGTTPSPVTQP 215
Query 68 LEEKRTGPVETPVQVGPLTTPTPEEEAESTEGEELAGVDSPSPLTEPLEEKRTGPV 123
+ + + PV P + PE +A L+G +PSP+T+P + + P+
Sbjct 216 MSQVQAAPVPEP-------SMNPEMQALL---RLLSGGTTPSPVTQPTSQVQAAPI 261
> 9258.ENSOANP00000018455
Length=410
Score = 31.6 bits (70), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 37/118 (31%), Gaps = 15/118 (12%)
Query 28 PVETPVQVGPVTEPAPEEEAESTEDEELVGVDGPSPPTLPLEEKRTGPVETPVQ---VGP 84
P P GP PA E A G GP P E GP TP GP
Sbjct 166 PTRDPGPAGPRASPAHEGSAGPGATPAHGGPAGPG--ATPAHEGPAGPGATPAHGGPAGP 223
Query 85 LTTP----------TPEEEAESTEGEELAGVDSPSPLTEPLEEKRTGPVETPVQVGPL 132
TP +P E + G A P P P E GP TP GP
Sbjct 224 GATPAHEGPAGPRASPAHEGSAGPGATPAHGGPPGPRATPAHEGSAGPRATPAHEGPA 281
Lambda K H
0.300 0.125 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 30377245073
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40