bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_4494_orf3
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
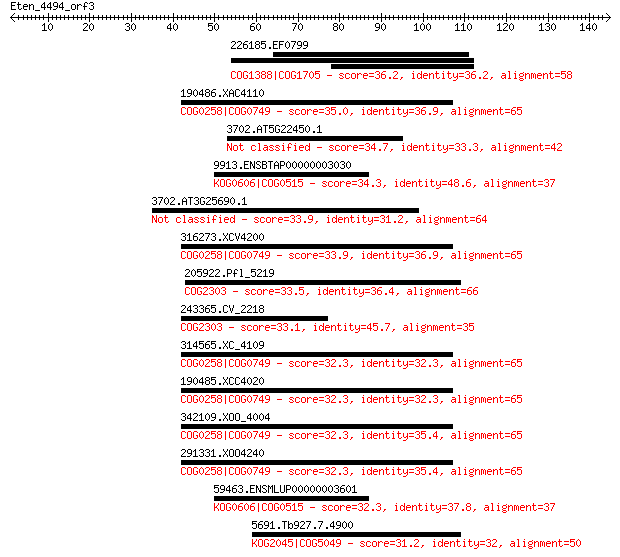
226185.EF0799 36.2 0.28
190486.XAC4110 35.0 0.60
3702.AT5G22450.1 34.7 0.79
9913.ENSBTAP00000003030 34.3 1.2
3702.AT3G25690.1 33.9 1.4
316273.XCV4200 33.9 1.5
205922.Pfl_5219 33.5 1.9
243365.CV_2218 33.1 2.4
314565.XC_4109 32.3 4.1
190485.XCC4020 32.3 4.1
342109.XOO_4004 32.3 4.4
291331.XOO4240 32.3 4.4
59463.ENSMLUP00000003601 32.3 4.5
5691.Tb927.7.4900 31.2 8.1
> 226185.EF0799
Length=737
Score = 36.2 bits (82), Expect = 0.28, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query 64 PGTPGPNSLSSSQA---LRSSAALNSVVAPFNITVANISAWLQLSGILLL 110
PGT G N+ S + ++S LN + A + ++VAN+ +W +SG L+
Sbjct 348 PGTGGSNNQSGTNTYYTVKSGDTLNKIAAQYGVSVANLRSWNGISGDLIF 397
Score = 35.0 bits (79), Expect = 0.68, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 54 GETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAWLQLSGILLLG 111
G ST+T T ++S ++S LN + A F ++VAN+ +W + G L+
Sbjct 611 GANSGSTNTNKPTNNGGGATTSYTIKSGDTLNKISAQFGVSVANLRSWNGIKGDLIFA 668
Score = 33.1 bits (74), Expect = 2.5, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 78 LRSSAALNSVVAPFNITVANISAWLQLSGILLLG 111
++S LN + A F ++VAN+ AW +SG L+
Sbjct 569 IKSGDTLNKISAQFGVSVANLQAWNNISGSLIFA 602
> 190486.XAC4110
Length=934
Score = 35.0 bits (79), Expect = 0.60, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
GAADG+ V G+ DF+ PG N++S S+ + S AA V+A F + I
Sbjct 119 GAADGLAVTISTGDKDFAQLVRPGIELVNTMSGSR-MDSDAA---VIAKFGVRPEQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 3702.AT5G22450.1
Length=1180
Score = 34.7 bits (78), Expect = 0.79, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 53 FGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNIT 94
F +DFS +PGT GP S+ S L+ L + +P+ ++
Sbjct 431 FATSDFSPRASPGTTGPLSVVDSSPLKMKRELRNASSPYGLS 472
> 9913.ENSBTAP00000003030
Length=1307
Score = 34.3 bits (77), Expect = 1.2, Method: Composition-based stats.
Identities = 18/37 (48%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 50 IFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNS 86
IF GE D TTP P P+SLS+ A S A L S
Sbjct 815 IFILGEPDAPPETTPVIPKPSSLSADTAALSHARLRS 851
> 3702.AT3G25690.1
Length=1004
Score = 33.9 bits (76), Expect = 1.4, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query 35 PLQVMVCGAADGICVIFCFGETDFSTSTTPGTPG-PNSLSSSQALRSSAALNSVVAPFNI 93
PL+ ++ A I FG+ D + TP TP P + QA LNSV A F++
Sbjct 486 PLESLMIRNAGESVAITTFGQVDQESPGTPETPNLPRIRTQQQASSPGEGLNSVAASFHV 545
Query 94 TVANI 98
++
Sbjct 546 MSKSV 550
> 316273.XCV4200
Length=933
Score = 33.9 bits (76), Expect = 1.5, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
GAADG+ V G+ DF+ PG N++S S+ + S AA V+A F + I
Sbjct 119 GAADGLQVTISTGDKDFAQLVRPGIELVNTMSGSR-MDSDAA---VIAKFGVRPDQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 205922.Pfl_5219
Length=1150
Score = 33.5 bits (75), Expect = 1.9, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 31/66 (46%), Gaps = 9/66 (13%)
Query 43 AADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAWL 102
A D CV+ C+G T F S G G S+ SQ A ++VVA T + A L
Sbjct 913 AKDVQCVVHCYGATTFFMSLLAGLQGVRSVVCSQ-----IAADTVVA----TATGLKAGL 963
Query 103 QLSGIL 108
L G+L
Sbjct 964 HLPGML 969
> 243365.CV_2218
Length=1151
Score = 33.1 bits (74), Expect = 2.4, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQ 76
GAAD CV+ C+G T F S G G S+ SQ
Sbjct 913 GAADVQCVVHCYGATTFFMSMLAGLEGVRSVVCSQ 947
> 314565.XC_4109
Length=927
Score = 32.3 bits (72), Expect = 4.1, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 30/65 (46%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
GA+DG+ V G+ DF+ PG N++S S+ A V+A F + I
Sbjct 119 GASDGLAVTISTGDKDFAQLVRPGVELVNTMSGSRMDSDEA----VIAKFGVRPNQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 190485.XCC4020
Length=927
Score = 32.3 bits (72), Expect = 4.1, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 30/65 (46%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
GA+DG+ V G+ DF+ PG N++S S+ A V+A F + I
Sbjct 119 GASDGLAVTISTGDKDFAQLVRPGVELVNTMSGSRMDSDEA----VIAKFGVRPNQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 342109.XOO_4004
Length=933
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 32/65 (49%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
AADG+ V G+ DF+ PG N++S S+ + S AA V+A F + I
Sbjct 119 AAADGLSVTISTGDKDFAQLVRPGIELVNTMSGSR-MDSDAA---VIAKFGVRPDQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 291331.XOO4240
Length=933
Score = 32.3 bits (72), Expect = 4.4, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 32/65 (49%), Gaps = 4/65 (6%)
Query 42 GAADGICVIFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAW 101
AADG+ V G+ DF+ PG N++S S+ + S AA V+A F + I
Sbjct 119 AAADGLSVTISTGDKDFAQLVRPGIELVNTMSGSR-MDSDAA---VIAKFGVRPDQIVDL 174
Query 102 LQLSG 106
L L G
Sbjct 175 LALMG 179
> 59463.ENSMLUP00000003601
Length=1128
Score = 32.3 bits (72), Expect = 4.5, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 50 IFCFGETDFSTSTTPGTPGPNSLSSSQALRSSAALNS 86
+F GE D + TPG P P+SLS+ L S
Sbjct 639 VFILGEPDLPPAATPGAPKPSSLSADPTALGQTRLRS 675
> 5691.Tb927.7.4900
Length=1418
Score = 31.2 bits (69), Expect = 8.1, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 59 STSTTPGTPGPNSLSSSQALRSSAALNSVVAPFNITVANISAWLQLSGIL 108
ST TP P P S + AA +VV+P ++TV+ S +Q++ ++
Sbjct 1252 STEGTPSIPKPRSAMGETSASGIAAGRNVVSPSSLTVSGGSGGIQVAELM 1301
Lambda K H
0.325 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22567178795
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40