bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
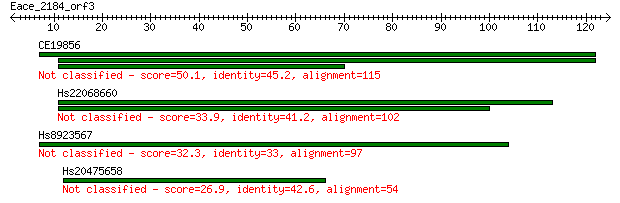
Query= Eace_2184_orf3
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
CE19856 50.1 1e-06
Hs22068660 33.9 0.085
Hs8923567 32.3 0.20
Hs20475658 26.9 9.6
> CE19856
Length=933
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 52/122 (42%), Positives = 52/122 (42%), Gaps = 22/122 (18%)
Query 7 PESVGGRVSGSNGGRVPGPTGV-VPGSAG-----GRVPDLNGGRVPGTTGV-VPGPVGGR 59
PE G RV G G RVPG G VPG RVP G RVPG G PG G R
Sbjct 482 PEFQGFRVPGFQGFRVPGFQGSRVPGIQSFRVQVSRVPGFQGARVPGIQGSRFPGIQGAR 541
Query 60 LPGPNGGRVSEPVGGVVPGPVGGRVSWSNGGPAPGPTGVVPEPVGGRVSGPNGGRVPGAT 119
G G R SE G VPG G RV PG G RV G G RVPG
Sbjct 542 FSGFQGSRDSEFQGSRVPGFQGARV--------PGIQGF-------RVPGFQGSRVPGFQ 586
Query 120 GV 121
G
Sbjct 587 GF 588
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 49/117 (41%), Positives = 49/117 (41%), Gaps = 24/117 (20%)
Query 11 GGRVSGSNGGRVPGPTGV-VPGSAGGRVPDLNGGRVPGTTGVVPGPV-----GGRLPGPN 64
G RV G G RVPG G VPG G RV RVPG G PG G R PG
Sbjct 563 GARVPGIQGFRVPGFQGSRVPGFQGFRVQ---VSRVPGFQG--PGFQGSRVSGSRFPGFQ 617
Query 65 GGRVSEPVGGVVPGPVGGRVSWSNGGPAPGPTGVVPEPVGGRVSGPNGGRVPGATGV 121
G RV VPG G RV G PG G RV G G RVPG G
Sbjct 618 GFRVQ---VSRVPGFQGFRVL---GSRFPGFQGF-------RVPGFQGARVPGIQGF 661
Score = 30.8 bits (68), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 30/75 (40%), Gaps = 18/75 (24%)
Query 11 GGRVSGSNG----------GRVPGPTGV------VPGSAGGRVPDLNGGRVPGTTGVVPG 54
G RVSGS RVPG G PG G RVP G RVPG G
Sbjct 605 GSRVSGSRFPGFQGFRVQVSRVPGFQGFRVLGSRFPGFQGFRVPGFQGARVPGIQGFRVQ 664
Query 55 PVGGRLPGPNGGRVS 69
R+ G G RVS
Sbjct 665 V--SRVTGFQGFRVS 677
> Hs22068660
Length=666
Score = 33.9 bits (76), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 42/105 (40%), Positives = 46/105 (43%), Gaps = 3/105 (2%)
Query 11 GGRVSGSNGGRVPGPTGV-VPGSAGGRVPDLNGGRVPGTTGV-VPGPVGGRLPGPNGGRV 68
G V G G VPG G VPG G VP + VPG G VPG G +PG G V
Sbjct 258 GAEVPGLEGAEVPGVEGAEVPGLEGAEVPGVEDAEVPGLEGAEVPGVEGAEVPGLEGAEV 317
Query 69 SEPVGGVVPGPVGGRVSWSNGGPAPGPTGV-VPEPVGGRVSGPNG 112
G VPG G V G PG G VP+ G V G +G
Sbjct 318 PGVEGAEVPGVEGAEVPGVEGAEVPGVKGAEVPDVEGAEVPGGHG 362
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 37/91 (40%), Positives = 39/91 (42%), Gaps = 2/91 (2%)
Query 11 GGRVSGSNGGRVPG-PTGVVPGSAGGRVPDLNGGRVPGTTGV-VPGPVGGRLPGPNGGRV 68
G V G G VPG VPG G VP + G VPG G VPG G +PG G V
Sbjct 274 GAEVPGLEGAEVPGVEDAEVPGLEGAEVPGVEGAEVPGLEGAEVPGVEGAEVPGVEGAEV 333
Query 69 SEPVGGVVPGPVGGRVSWSNGGPAPGPTGVV 99
G VPG G V G PG G V
Sbjct 334 PGVEGAEVPGVKGAEVPDVEGAEVPGGHGWV 364
> Hs8923567
Length=146
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 41/105 (39%), Gaps = 14/105 (13%)
Query 7 PESVGGRVSGSNGGRVPGPTGVVPGSAGGRVPDLNGGRVPGTTGV-------VPGP-VGG 58
P + G V S R PGP G+ GR+ G G P P +
Sbjct 17 PATEAGSVQRSLWFRFPGPQN---GTQHRFCSRSRSGRMEGAKGKECTAGHSWPAPALWC 73
Query 59 RLPGPNGGRVSEPVGGVVPGPVGGRVSWSNGGPAPGPTGVVPEPV 103
LP + GR +EP G+V P R W P PG +VP P+
Sbjct 74 LLPVTSEGRWTEPPSGLVLFPATERAGWH---PTPGEQYLVPHPL 115
> Hs20475658
Length=186
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 12 GRVSGSNGGRVPGPTGVVPGSAGGRVPDLNGGRVP-GTTGVVPGPVG---GRLPGPNG 65
GRV + PT ++P S G PD+ G +P G GV P G GRLP P+G
Sbjct 99 GRVECTTAT---APTCILPTSQAGANPDVQDGSLPAGGAGVNPATQGTPAGRLPTPSG 153
Lambda K H
0.309 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40