bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2545_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
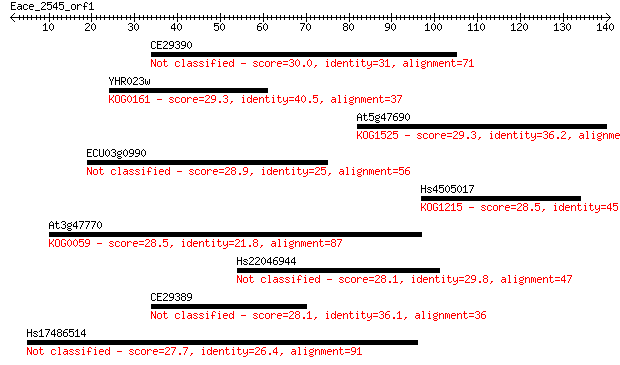
CE29390 30.0 1.4
YHR023w 29.3 2.5
At5g47690 29.3 2.7
ECU03g0990 28.9 2.9
Hs4505017 28.5 3.7
At3g47770 28.5 4.8
Hs22046944 28.1 4.9
CE29389 28.1 5.0
Hs17486514 27.7 7.5
> CE29390
Length=586
Score = 30.0 bits (66), Expect = 1.4, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 2/71 (2%)
Query 34 AKRNLDETNTLCGRLMVSIEAQCRGVQIRREWSSAYGKHFSVATSQAPKVGSLVQEFTEL 93
A + ++ N C L V+ +A C RR WS+AY T V V+ E
Sbjct 474 APKLTNKFNKCCHNLFVTDDACCPQAAARRYWSTAYEVCMPTVTVDFSPVRFEVKFDDE- 532
Query 94 KRRGDQVYFTD 104
RG +V+FT+
Sbjct 533 -NRGRKVHFTE 542
> YHR023w
Length=1928
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query 24 NKELEEKIKFAKRNLD---ETNTLCGRLMVSIEAQCRGVQ 60
NKE+ EKIK+ + L E N+ G L+ +++A C G +
Sbjct 1712 NKEITEKIKYLEETLQLQMEQNSRNGELVKTLQASCNGYK 1751
> At5g47690
Length=1638
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 82 KVGSLVQEFTELKRRGDQVYFTDILLSDANVTAKKLEEDVVVLQLPVLEGLKDCLFSD 139
KV LV E L R F I L K+L + VV +++ +L+ +KDCL SD
Sbjct 284 KVVGLVGELFSLPGRVISEEFDSIFLE----FLKRLTDRVVEVRMAILDHIKDCLLSD 337
> ECU03g0990
Length=569
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/56 (25%), Positives = 25/56 (44%), Gaps = 0/56 (0%)
Query 19 LMRSANKELEEKIKFAKRNLDETNTLCGRLMVSIEAQCRGVQIRREWSSAYGKHFS 74
+ + N + +E +K ++ + N + G + S A C G R W +GK S
Sbjct 88 IAKQENDDYKEVVKLWMKSTEHRNNILGDYVYSGVATCVGKDGNRYWVQVFGKDVS 143
> Hs4505017
Length=1613
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 97 GDQVYFTDILL-SDANVTAKKLEEDVVVLQLPVLEGLK 133
GD VY+TD S V + E +V++ QLP L GLK
Sbjct 542 GDYVYWTDWQRRSIERVHKRSAEREVIIDQLPDLMGLK 579
> At3g47770
Length=722
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 19/91 (20%), Positives = 43/91 (47%), Gaps = 6/91 (6%)
Query 10 PGANSSVSLLMRSANKELEEKIKFAKRNLDETNTLCGRLMVSIEAQCRGVQIRREWSSAY 69
P + ++ +++ A K I +++E LC RL + ++ + + + +E Y
Sbjct 577 PASRINLWTVIKRAKKH--AAIILTTHSMEEAEFLCDRLGIFVDGRLQCIGNPKELKGRY 634
Query 70 GKHFSVATSQAPK----VGSLVQEFTELKRR 96
G + + + +P+ V +LVQE + R+
Sbjct 635 GGSYVLTITTSPEHEKDVETLVQEVSSNARK 665
> Hs22046944
Length=443
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 54 AQCRGVQIRREWSSAYGKHFSVATSQAPKVGSLVQEFTELKRRGDQV 100
AQ G ++ + W + +V T+Q+ +VG+ TEL+ GD +
Sbjct 159 AQKNGEELDKHWPQQIEESTTVVTTQSAEVGAAETTLTELRLLGDDL 205
> CE29389
Length=575
Score = 28.1 bits (61), Expect = 5.0, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 34 AKRNLDETNTLCGRLMVSIEAQCRGVQIRREWSSAY 69
A + ++ N C L V+ +A C RR WS+AY
Sbjct 474 APKLTNKFNKCCHNLFVTDDACCPQAAARRYWSTAY 509
> Hs17486514
Length=517
Score = 27.7 bits (60), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 24/91 (26%), Positives = 41/91 (45%), Gaps = 3/91 (3%)
Query 5 GKRKAPGANSSVSLLMRSANKELEEKIKFAKRNLDETNTLCGRLMVSIEAQCRGVQIRRE 64
G RK S L + + + L+E++ F K + +ET+T + AQ ++
Sbjct 338 GLRKLTDDTSVTWLQLETEIEALKEELLFTKNHEEETDTWAQYDKL---AQKNPEELDMY 394
Query 65 WSSAYGKHFSVATSQAPKVGSLVQEFTELKR 95
WS + +V T QA ++G TEL+
Sbjct 395 WSQQIEESTTVVTKQAAEIGVAEMILTELRH 425
Lambda K H
0.315 0.130 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40