bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4270_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
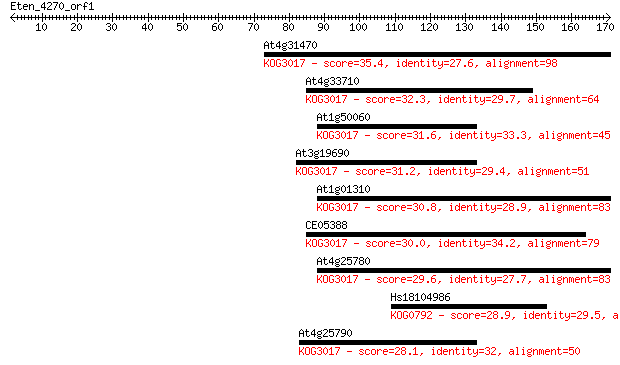
At4g31470 35.4 0.050
At4g33710 32.3 0.51
At1g50060 31.6 0.86
At3g19690 31.2 0.92
At1g01310 30.8 1.2
CE05388 30.0 2.1
At4g25780 29.6 2.6
Hs18104986 28.9 5.7
At4g25790 28.1 9.1
> At4g31470
Length=185
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 49/101 (48%), Gaps = 17/101 (16%)
Query 73 VVRQER-MTMDKFREVMLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEH 131
+ Q++ + + ++ L HN +R + LP LKW++ LA ++ + + +C++ H
Sbjct 38 IFSQDKALARNTIQQQFLRPHNILRAKLRLPPLKWSNSLALYASRWARTRR--GDCKLIH 95
Query 132 S--PHEARELPNMKPGIGENLWTGCTVEPLPTDIPSSWASE 170
S P+ GENL+ G P D ++WASE
Sbjct 96 SGGPY------------GENLFWGSGKGWTPRDAVAAWASE 124
> At4g33710
Length=166
Score = 32.3 bits (72), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query 85 REVMLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHSPHEARELPNMKP 144
R+ L+ HN R S+P +KW+ A Y Q + ++CR+ HS R N+
Sbjct 31 RQDYLDVHNHARDDVSVPHIKWHAGAARYAWNYAQRRK--RDCRLIHSNSRGRYGENLAW 88
Query 145 GIGE 148
G+
Sbjct 89 SSGD 92
> At1g50060
Length=161
Score = 31.6 bits (70), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Query 88 MLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHS 132
L HN R Q +P + W+ LAA L Y + +C + HS
Sbjct 30 YLNSHNTARAQVGVPNVVWDTTLAAYALNYSNFRK--ADCNLVHS 72
> At3g19690
Length=161
Score = 31.2 bits (69), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 82 DKFREVMLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHS 132
+ ++ LE HN R + L L W+D +AA Y + + +C + HS
Sbjct 23 EDLQQQFLEAHNEARNEVGLDPLVWDDEVAAYAASYANQR--INDCALVHS 71
> At1g01310
Length=294
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 16/85 (18%)
Query 88 MLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHS--PHEARELPNMKPG 145
L HN VR + P +W+ LAA + + + +CR+ HS P+
Sbjct 141 FLIAHNLVRARVGEPPFQWDGRLAAYARTWANQR--VGDCRLVHSNGPY----------- 187
Query 146 IGENLWTGCTVEPLPTDIPSSWASE 170
GEN++ P DI + WA E
Sbjct 188 -GENIFWAGKNNWSPRDIVNVWADE 211
> CE05388
Length=209
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 40/91 (43%), Gaps = 24/91 (26%)
Query 85 REVMLEGHNAVRR-----QHSLPGLKWNDLLAANMLKY-------LQHQNLLQECRMEHS 132
++ M+ HNAVR ++ G K + A NMLK QN C M+HS
Sbjct 23 KQSMVNAHNAVRSSIAKGEYVAKGTKKDS--ATNMLKMKWDNSLAQSAQNYANGCPMQHS 80
Query 133 PHEARELPNMKPGIGENLWTGCTVEPLPTDI 163
P ++ GENL+ + P+ TD+
Sbjct 81 PDKS---------YGENLFWAYSSSPI-TDL 101
> At4g25780
Length=190
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 8/83 (9%)
Query 88 MLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHSPHEARELPNMKPGIG 147
L HN VR P L W+ L + + +C + HS + N + +G
Sbjct 55 FLFRHNLVRAARFEPPLIWDRRLQNYAQGWANQRR--GDCALRHS------VSNGEFNLG 106
Query 148 ENLWTGCTVEPLPTDIPSSWASE 170
EN++ G P D +WASE
Sbjct 107 ENIYWGYGANWSPADAVVAWASE 129
> Hs18104986
Length=913
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 109 LLAANMLKYLQHQNLLQECRMEHSPHEARELPNMKPGIGENLWT 152
++A NML Y +NL + C H+ +A++L + + WT
Sbjct 283 IVAFNMLNYRSCKNLWKSCVEHHTFFQAKKLLPQEKNVLSQYWT 326
> At4g25790
Length=210
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query 83 KFREVMLEGHNAVRRQHSLPGLKWNDLLAANMLKYLQHQNLLQECRMEHS 132
F + L+ HN VR LP L W D+ A+ + +Q +C + HS
Sbjct 73 SFEQQFLDPHNTVRGGLGLPPLVW-DVKIASYATWWANQRRY-DCSLTHS 120
Lambda K H
0.318 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2490594054
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40