bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6775_orf1
Length=131
Score E
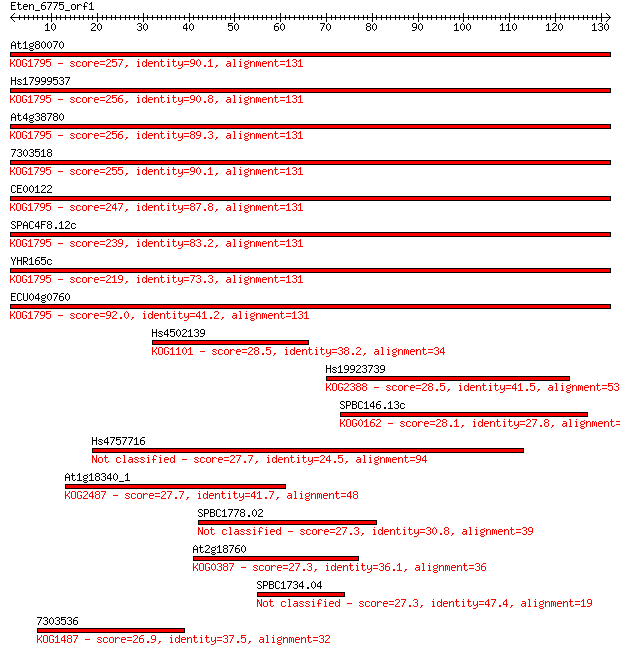
Sequences producing significant alignments: (Bits) Value
At1g80070 257 4e-69
Hs17999537 256 6e-69
At4g38780 256 1e-68
7303518 255 2e-68
CE00122 247 4e-66
SPAC4F8.12c 239 7e-64
YHR165c 219 1e-57
ECU04g0760 92.0 2e-19
Hs4502139 28.5 3.2
Hs19923739 28.5 3.7
SPBC146.13c 28.1 4.6
Hs4757716 27.7 5.8
At1g18340_1 27.7 6.6
SPBC1778.02 27.3 6.9
At2g18760 27.3 8.0
SPBC1734.04 27.3 8.4
7303536 26.9 8.7
> At1g80070
Length=2382
Score = 257 bits (657), Expect = 4e-69, Method: Compositional matrix adjust.
Identities = 118/131 (90%), Positives = 127/131 (96%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMPSRFPPV+FYTPKE+GGLGMLSMGHILIPQSDLRYS+QT+ G+THFRSGMSHEED
Sbjct 1350 LNSKMPSRFPPVIFYTPKEIGGLGMLSMGHILIPQSDLRYSKQTDVGVTHFRSGMSHEED 1409
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
QLIPNLYRY+Q WESEF++SQRVWAEYALKR EA AQ+RRLTLEDLEDSWDRGIPRINTL
Sbjct 1410 QLIPNLYRYIQPWESEFIDSQRVWAEYALKRQEAQAQNRRLTLEDLEDSWDRGIPRINTL 1469
Query 121 FQKDRHTLAYD 131
FQKDRHTLAYD
Sbjct 1470 FQKDRHTLAYD 1480
> Hs17999537
Length=2335
Score = 256 bits (655), Expect = 6e-69, Method: Compositional matrix adjust.
Identities = 119/131 (90%), Positives = 127/131 (96%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMPSRFPPVVFYTPKELGGLGMLSMGH+LIPQSDLR+S+QT+ GITHFRSGMSHEED
Sbjct 1303 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHVLIPQSDLRWSKQTDVGITHFRSGMSHEED 1362
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
QLIPNLYRY+Q WESEF++SQRVWAEYALKR EA AQ+RRLTLEDLEDSWDRGIPRINTL
Sbjct 1363 QLIPNLYRYIQPWESEFIDSQRVWAEYALKRQEAIAQNRRLTLEDLEDSWDRGIPRINTL 1422
Query 121 FQKDRHTLAYD 131
FQKDRHTLAYD
Sbjct 1423 FQKDRHTLAYD 1433
> At4g38780
Length=2352
Score = 256 bits (653), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 117/131 (89%), Positives = 126/131 (96%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMPSRFPPV+FYTPKE+GGLGMLSMGHILIPQSDLRYS QT+ G++HFRSGMSHEED
Sbjct 1322 LNSKMPSRFPPVIFYTPKEIGGLGMLSMGHILIPQSDLRYSNQTDVGVSHFRSGMSHEED 1381
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
QLIPNLYRY+Q WESEF++SQRVWAEYALKR EA AQ+RRLTLEDLEDSWDRGIPRINTL
Sbjct 1382 QLIPNLYRYIQPWESEFIDSQRVWAEYALKRQEAQAQNRRLTLEDLEDSWDRGIPRINTL 1441
Query 121 FQKDRHTLAYD 131
FQKDRHTLAYD
Sbjct 1442 FQKDRHTLAYD 1452
> 7303518
Length=2396
Score = 255 bits (651), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 118/131 (90%), Positives = 127/131 (96%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMPSRFPPVVFYTPKELGGLGMLSMGH+LIPQSDLR+S+QT+ GITHFRSGMSH+ED
Sbjct 1363 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHVLIPQSDLRWSKQTDVGITHFRSGMSHDED 1422
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
QLIPNLYRY+Q WESEF++SQRVWAEYALKR EA AQ+RRLTLEDLEDSWDRGIPRINTL
Sbjct 1423 QLIPNLYRYIQPWESEFIDSQRVWAEYALKRQEANAQNRRLTLEDLEDSWDRGIPRINTL 1482
Query 121 FQKDRHTLAYD 131
FQKDRHTLAYD
Sbjct 1483 FQKDRHTLAYD 1493
> CE00122
Length=2329
Score = 247 bits (630), Expect = 4e-66, Method: Compositional matrix adjust.
Identities = 115/132 (87%), Positives = 126/132 (95%), Gaps = 1/132 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESG-ITHFRSGMSHEE 59
LNSKMPSRFPPVVFYTPKE+GGLGMLSMGH+LIPQSDLR+ QQTE+G +THFRSGMSH+E
Sbjct 1295 LNSKMPSRFPPVVFYTPKEIGGLGMLSMGHVLIPQSDLRWMQQTEAGGVTHFRSGMSHDE 1354
Query 60 DQLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINT 119
DQLIPNLYRY+Q WE+EFV+S RVWAEYALKR EA AQ+RRLTLEDL+DSWDRGIPRINT
Sbjct 1355 DQLIPNLYRYIQPWEAEFVDSVRVWAEYALKRQEANAQNRRLTLEDLDDSWDRGIPRINT 1414
Query 120 LFQKDRHTLAYD 131
LFQKDRHTLAYD
Sbjct 1415 LFQKDRHTLAYD 1426
> SPAC4F8.12c
Length=2363
Score = 239 bits (611), Expect = 7e-64, Method: Compositional matrix adjust.
Identities = 109/131 (83%), Positives = 122/131 (93%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMPSRFPP VFY+PKELGGLGMLSMGH+LIPQSDLR+S+QT++GITHFRSGM+ +
Sbjct 1327 LNSKMPSRFPPAVFYSPKELGGLGMLSMGHVLIPQSDLRWSKQTDTGITHFRSGMTTNGE 1386
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
LIPNLYRY+Q WESEF++SQRVWAEYA+KR EA Q+RRLTLEDLEDSWDRGIPRINTL
Sbjct 1387 HLIPNLYRYIQPWESEFIDSQRVWAEYAMKRQEALQQNRRLTLEDLEDSWDRGIPRINTL 1446
Query 121 FQKDRHTLAYD 131
FQKDRHTLAYD
Sbjct 1447 FQKDRHTLAYD 1457
> YHR165c
Length=2413
Score = 219 bits (557), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 96/131 (73%), Positives = 117/131 (89%), Gaps = 0/131 (0%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
LNSKMP+RFPP VFYTPKELGGLGM+S HILIP SDL +S+QT++GITHFR+GM+HE++
Sbjct 1375 LNSKMPTRFPPAVFYTPKELGGLGMISASHILIPASDLSWSKQTDTGITHFRAGMTHEDE 1434
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
+LIP ++RY+ TWE+EF++SQRVWAEYA KR EA Q+RRL E+LE SWDRGIPRI+TL
Sbjct 1435 KLIPTIFRYITTWENEFLDSQRVWAEYATKRQEAIQQNRRLAFEELEGSWDRGIPRISTL 1494
Query 121 FQKDRHTLAYD 131
FQ+DRHTLAYD
Sbjct 1495 FQRDRHTLAYD 1505
> ECU04g0760
Length=2172
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 69/131 (52%), Gaps = 30/131 (22%)
Query 1 LNSKMPSRFPPVVFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEED 60
+NSKMP RFPPV+FY PKE+GGLGMLS+G I + D S+ D
Sbjct 1299 INSKMPVRFPPVMFYAPKEMGGLGMLSVGGIRVSHEDFEGSEDV--------------ND 1344
Query 61 QLIPNLYRYLQTWESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTL 120
++IP Y+ W+ EF ES RVW EY R +E D+GIPR++TL
Sbjct 1345 RMIPAAMDYVSRWDYEFEESNRVWKEYG----------RSGKMEP-----DKGIPRMSTL 1389
Query 121 FQKDRHTLAYD 131
Q+ R L YD
Sbjct 1390 LQRSRWLL-YD 1399
> Hs4502139
Length=604
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 32 LIPQSDLRYSQQTESGITHFRSGMSHEEDQLIPN 65
L+ SD + ES I HF G H ED ++ N
Sbjct 345 LLSTSDSPGDENAESSIIHFEPGEDHSEDAIMMN 378
> Hs19923739
Length=505
Score = 28.5 bits (62), Expect = 3.7, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 6/55 (10%)
Query 70 LQTWESE--FVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTLFQ 122
LQ WESE F SQ A L A Q RL + + +D G+P TLFQ
Sbjct 87 LQAWESEGLFQISQNKVAVLLL----AGGQGTRLGVAYPKGMYDVGLPSRKTLFQ 137
> SPBC146.13c
Length=1217
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 27/54 (50%), Gaps = 10/54 (18%)
Query 73 WESEFVESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDRGIPRINTLFQKDRH 126
W++ QR W Y +R+EAAA ++L W+R ++N ++ R+
Sbjct 723 WDTMATRIQRAWRSYVRRRSEAAACIQKL--------WNRN--KVNMELERVRN 766
> Hs4757716
Length=293
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 40/94 (42%), Gaps = 13/94 (13%)
Query 19 ELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRSGMSHEEDQLIPNLYRYLQTWESEFV 78
E+ GL S G D RY + F+ + +++L + YR + W+ +
Sbjct 86 EVAGLTSFSFGE----DDDCRY-------VMIFKKEFAPSDEEL--DSYRRGEEWDPQKA 132
Query 79 ESQRVWAEYALKRAEAAAQSRRLTLEDLEDSWDR 112
E +R E A ++ E AAQ + + D D+
Sbjct 133 EEKRKLKELAQRQEEEAAQQGPVVVSPASDYKDK 166
> At1g18340_1
Length=289
Score = 27.7 bits (60), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Query 13 VFYTPKELGGLGMLSMGHILIPQSDLRYSQQTESGITHFRS--GMSHEED 60
V +TPK+L GL +L+PQ D RY S HF S G S +D
Sbjct 204 VHHTPKQLDGL--FQYLTVLLPQEDNRYGLHMLSVSVHFLSVFGQSKLDD 251
> SPBC1778.02
Length=693
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 42 QQTESGITHFRSGMSHEE-DQLIPNLYRYLQTWESEFVES 80
+ ++ ++ RS ++H E D+ I N+ RY + E +F+E+
Sbjct 633 HELQNEVSILRSSVNHREVDEAIDNILRYTNSTEQQFLEA 672
> At2g18760
Length=1187
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 41 SQQTESGITHFRSGMSHEEDQLIPNLYRYLQTWESE 76
S T S + HFR +S + QL NL + + T E +
Sbjct 1138 SADTTSIVNHFRDIVSFNDKQLFKNLLKEIATLEKD 1173
> SPBC1734.04
Length=304
Score = 27.3 bits (59), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 9/19 (47%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 55 MSHEEDQLIPNLYRYLQTW 73
M H++D ++PN+YR L W
Sbjct 236 MRHDKDIIVPNVYRPLPDW 254
> 7303536
Length=368
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 4/32 (12%)
Query 7 SRFPPVVFYTPKELGGLGMLSMGHILIPQSDL 38
++ PP ++Y K+ GG+ + SM +PQS+L
Sbjct 173 NKKPPNIYYKRKDKGGINLNSM----VPQSEL 200
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40