bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7665_orf2
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
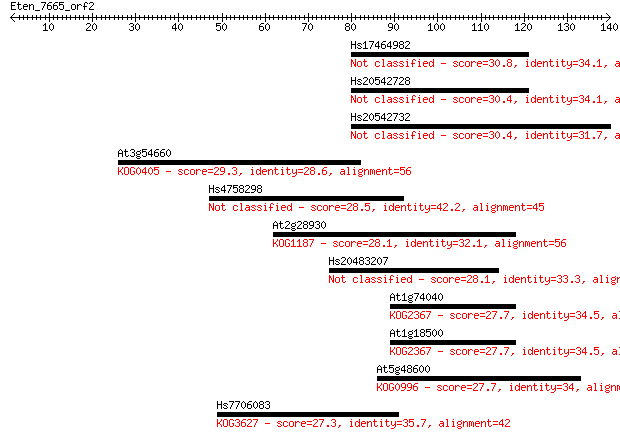
Hs17464982 30.8 0.74
Hs20542728 30.4 0.96
Hs20542732 30.4 1.1
At3g54660 29.3 2.3
Hs4758298 28.5 4.0
At2g28930 28.1 4.7
Hs20483207 28.1 4.8
At1g74040 27.7 6.7
At1g18500 27.7 7.1
At5g48600 27.7 7.5
Hs7706083 27.3 9.1
> Hs17464982
Length=318
Score = 30.8 bits (68), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 80 GVVSVASLLPQDAARAEQQALLSLLHEIEKSNSNTVKVSLR 120
G+ V ++P R EQ+ +LSL H + N + SLR
Sbjct 252 GIAMVVYMVPDSNQREEQEKMLSLFHSVFNPMLNPLIYSLR 292
> Hs20542728
Length=268
Score = 30.4 bits (67), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 80 GVVSVASLLPQDAARAEQQALLSLLHEIEKSNSNTVKVSLR 120
G+ V ++P R EQ+ +LSL H + N + SLR
Sbjct 210 GIAMVVYIVPDSNQREEQEKMLSLFHSVLNPILNPLIYSLR 250
> Hs20542732
Length=310
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 6/60 (10%)
Query 80 GVVSVASLLPQDAARAEQQALLSLLHEIEKSNSNTVKVSLRVFHLQQLAAIRGASYLRLQ 139
G+ V L+P ++ R +QQ +L+L + + N + SLR A ++GA Y LQ
Sbjct 252 GMAMVVYLVPDNSQRQKQQKILTLFYSLFNPLLNPLIYSLRN------AQVKGALYRALQ 305
> At3g54660
Length=565
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 2/56 (3%)
Query 26 QGCLPKNDDLFYFATRYVHSFSNLHRASHTTSCQGSRCWLHAAAQATAAVCPGTGV 81
+GC+PK L +A++Y H F + H + S W A A + TG+
Sbjct 138 RGCVPKK--LLVYASKYSHEFEDSHGFGWKYETEPSHDWTTLIANKNAELQRLTGI 191
> Hs4758298
Length=1255
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query 47 SNLHRASHTTS--CQGSRCWLHAAAQA---TAAVCPGTGVVSVASLLPQD 91
+N RA H S C+GSRCW ++ T VC G G LP D
Sbjct 186 TNRSRACHPCSPMCKGSRCWGESSEDCQSLTRTVCAG-GCARCKGPLPTD 234
> At2g28930
Length=412
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 62 RCWLHAAAQATAAVCPGTGVVSVASLLPQDAARAEQQALLSLLHEIEKSNSNTVKV 117
+ W+ Q A PGTGVV L QD + Q+ L + + + S+ N VK+
Sbjct 85 KGWIDE--QTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKL 138
> Hs20483207
Length=413
Score = 28.1 bits (61), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 18/39 (46%), Gaps = 0/39 (0%)
Query 75 VCPGTGVVSVASLLPQDAARAEQQALLSLLHEIEKSNSN 113
CP VV AS PQ E ++S +H + K + N
Sbjct 344 TCPVVTVVRAASWTPQGPHAWESGPVISFIHAVNKPSKN 382
> At1g74040
Length=631
Score = 27.7 bits (60), Expect = 6.7, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 89 PQDAARAEQQALLSLLHEIEKSNSNTVKV 117
P+DA R+E++ L +L E+ K+ + T+ +
Sbjct 231 PEDAGRSEREYLYEILGEVIKAGATTLNI 259
> At1g18500
Length=660
Score = 27.7 bits (60), Expect = 7.1, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 89 PQDAARAEQQALLSLLHEIEKSNSNTVKV 117
P+DA R+E++ L +L E+ K+ + T+ +
Sbjct 233 PEDAGRSEREYLYEILGEVIKAGATTLNI 261
> At5g48600
Length=1241
Score = 27.7 bits (60), Expect = 7.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 86 SLLPQDAARAEQQALLSLLHEIEKSNSNTVKVSLRVFHLQQLAAIRG 132
+L+PQ+ A E+ A L EKS + +K LR Q+ I G
Sbjct 513 TLVPQEQAAREKVAELKSAMNSEKSQNEVLKAVLRAKENNQIEGIYG 559
> Hs7706083
Length=487
Score = 27.3 bits (59), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 19/42 (45%), Gaps = 2/42 (4%)
Query 49 LHRASHTTSCQGSRCWLHAAAQATAAVCPGTGVVSVASLLPQ 90
L R+ H+ C G+ WL CP G V +A LPQ
Sbjct 11 LWRSPHSKGCPGAMWWL--LLWGVLQACPTRGSVLLAQELPQ 50
Lambda K H
0.320 0.128 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40